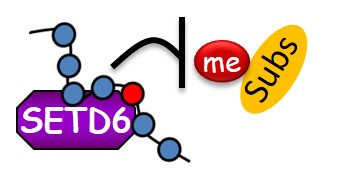
Lysine methylation signaling at Chromatin:
We are utilizing multi-disciplinary experimental platforms to unravel novel functional interplay between protein methylation of histones and non-histones and diverse signaling pathways ranging from cancer, metabolism to cell differentiation and immune response. We work mainly on the PKMTs SETD3 and SETD6 but not restricted to them. In recent years we were able to identify numerous new methylated substrates and interacting proteins for these enzymes and deciphered their mechanistic and physiological consequences in many pathological contexts .
Biochemical properties of Lysine methylation:
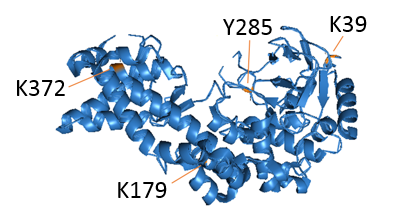
We study the biochemical properties of SETD6. We showed that the auto-methylation and the self-dimerization of SETD6 are important for its enzymatic activity. In a close collaboration with Prof. Amir Aharoni, BGU, we study basic mechanisms of enzyme-substrates recognition focusing on the PKMTs SETD6 and SETD7. To do so, we utilize classical biochemical and biophysical assays. In addition, we develop new strategies to follow the kinetics of methylation, both in-vitro and in cells .
Protein microarrays technologies:
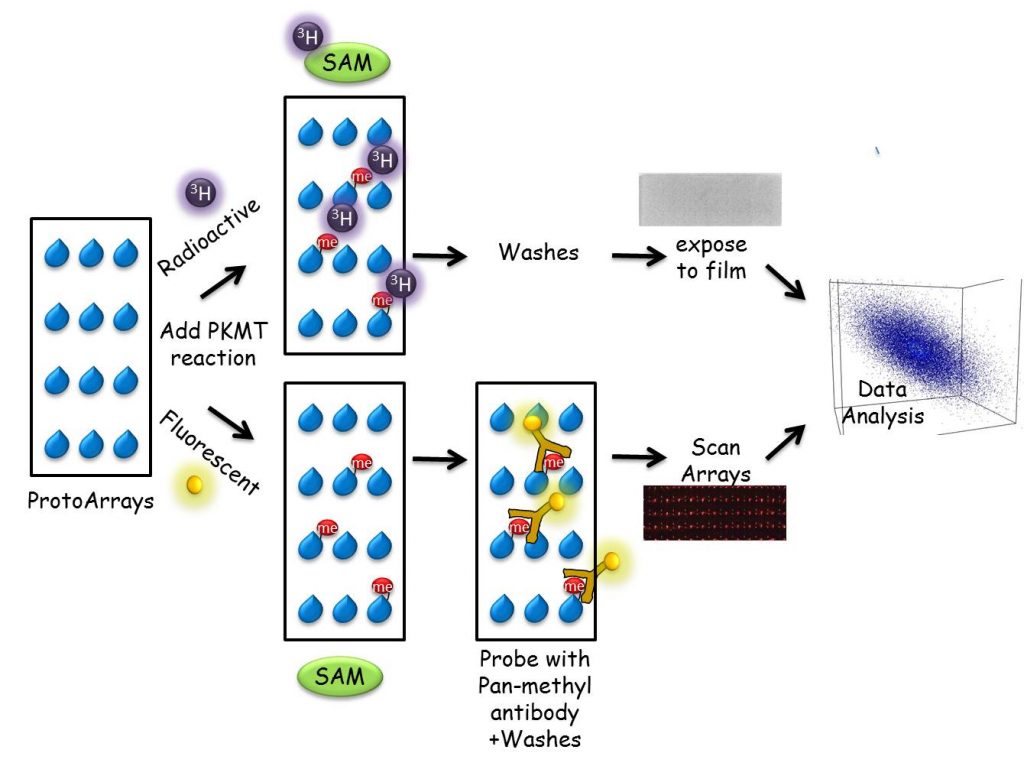
We have developed a unique proteomic methodology to identify new substrates for different PKMTs. Using a ProtoArray-based proteomic platform, we demonstrated that more than 9500 unique substrates can be screened in a single experiment in a reproducible and efficient manner (Refs). Employing this system, we identified numerous new targets for the PKMTs SETD6, SETD7 and SETD8. As part of our research program, we will continue to harness and develop protein and peptide array proteomic tools towards the goal of understanding how novel methylation signaling networks impact diverse cellular processes.
Applicative directions:
We aim to utilize our basic science findings and move toward a more applicative directions. We are performing small-molecule and rational-design peptide screens to modulate the enzymatic activity, substrate specificity, biochemical properties and cellular function of the different PKMTs.