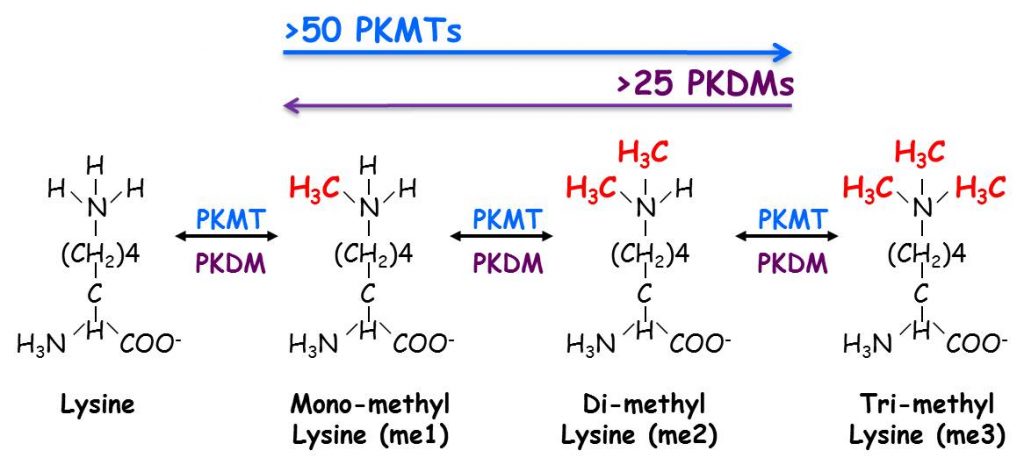
Biological systems respond to extracellular and intracellular signals in myriad ways by activating diverse signaling pathways. Reversible covalent post-translational modifications (PTMs) such as phosphorylation, acetylation and methylation are critical modulators of these signaling pathways. While phosphorylation is the most extensively studied PTM, lysine methylation is emerging as a key player in regulating intracellular signaling pathways. The methylation of lysine residues is performed by protein lysine (K) methyltransferases (PKMTs). It is estimated that in the human proteome there are ~50 PKMT and greater than 25 demethylases (PKDMs) which can reverse the process and remove the methyl mark away. A lysine residue can be mono, di or tri methylated.
The degree of methylation is enzyme specific and can define a distinct biological outcome by recruiting specialized regulatory factors named “readers” that specifically recognize distinct modification in a state (degree of methylation) and in a sequence dependent manner.