Have you ever wondered how do we know who are the microbes in environmental samples?
Understanding microbial life requires us to study their functionality and metabolism which is also only possible in pure culture. Sadly, with the increasing technological advances, isolating new microbes from the environment was pretty much abandoned, as it is not an easy task. Cultivating pure microorganisms is essential to form databases to which environmental samples will be compared to, however, some environments have more representatives than others: Most of the human and mice microbiome have representative isolates (Walker et al., 2014) however, for the rumen it is not the case. Only 117 cultures of rumen origin are available for purchase across several international culture collections.
It’s possible for you to contemplate writing on subsequent themes linked to science and engineering. Essay writing help on such topics should become an outstanding workout for the head and mind. It truly is going to ultimately allow you to come up plus a excellent research papers. Through this kind of composition writing activities, students might learn about various theories by getting in their own basics.
You’ve already been told your interest in and comprehension of the subject is essential that you have the capability to write your papers well. That’s only part of day to day creating practice. The overall amount of investigation you must do will change, dependant up on this issue. The creating requests you’re find under are in the likeness of questions.
The cause and impact issues aren’t restricted merely to subjects associated with scientific discipline. The facts are that composing editorials is among the most reliable strategies to generate awareness about health problems. There is plenty of ideas that you can come up with.
Luckily, The Hungate1000 project has aimed to increase whole genome sequencing data and culture collection and recently published 410 genomes of rumen microbes (Seshadri et al., 2018). Yet, both culture collection and the Hungate1000 collection do not represent the actual rumen composition, which makes our estimations about the rumen somewhat lacking.
Therefore, in our new paper Insights into the culturomics of the rumen microbiome we sought to determine the culturomics potential of the rumen microbiome and identify the factors that influence the cultivability of microbes from the rumen, without isolating them (Zehavi et al,. 2018, doi: 10.3389/fmicb.2018.01999).
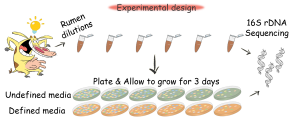
We managed to cultivate 23% of the rumen microbes and enrich for a large fraction of microbes which were not detected in the original rumen sample, or in the rumen of 38 additional cows. We hypothesize that this fraction belongs to the rumen rare biosphere, as they were enriched by our media, making them detectable. This finding implies that the rumen is probably far richer and more complex than we know, however, the role of this rare biosphere in the rumen is yet to be revealed.
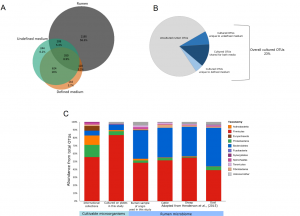
Additionally, using rumen fluid in the culturing media together with a defined media enabled us to increase the richness on the plates. Other factors that highly contributed for culturing were dilutions of the rumen sample and replications of the agar. Additionally, we identified that there are multifactorial genetic traits of microbes that contribute to their ability to be cultivated. Finally, the abundance of microbes in their original sample was highly correlated to the cultivability but, sample dilutions greatly contribute to removing this dependence.
Our point is, cultivating the yet uncultured microbes from the rumen is not a dream but a viable opportunity to better understand who is there, where are they doing and combine this knowledge with past and future studies.
References:
Walker, A. W., S. H. Duncan, P. Louis and H. J. Flint (2014). Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends in microbiology 22(5): 267-274.
Seshadri, Rekha, Sinead C. Leahy, Graeme T. Attwood, Koon Hoong Teh, Suzanne C. Lambie, Adrian L. Cookson, Emiley A. Eloe-Fadrosh, et al. (2018). Cultivation and Sequencing of Rumen Microbiome Members from the Hungate1000 Collection. Nature Biotechnology 36, 359–67.