What We Do?
Our Research
Our main research interest is to understand the ecological and evolutionary forces that shape microbial communities in nature and specifically, in gut environments. Understanding these forces enables us to predict and modulate the composition of the microbiome towards optimized functionality.
Microbial communities are everywhere and drive many basic processes in our everyday life, concerning agriculture, health and the environment. We study the evolutionary forces that act on microbial communities in nature, as well as the ecological forces that shape them. Specifically, we are interested in the factors that determine microbial community assembly in gut environments, as well as the effect of gene mobility via plasmids on their ecology.
For this purpose, we use state of the art tools, such as deep sequencing, big data analysis, metabolomics, fluorescence-activated cell sorting (FACS), high-performance microscopy and classical aerobic and anaerobic microbiology approaches. This knowledge is, in turn, used to understand and to influence the role of microbial communities in health and disease, as well as in agricultural systems. Hence, it is expected to have implications for future developments in food sustainability, renewable energy and medicine.
ongoing projects
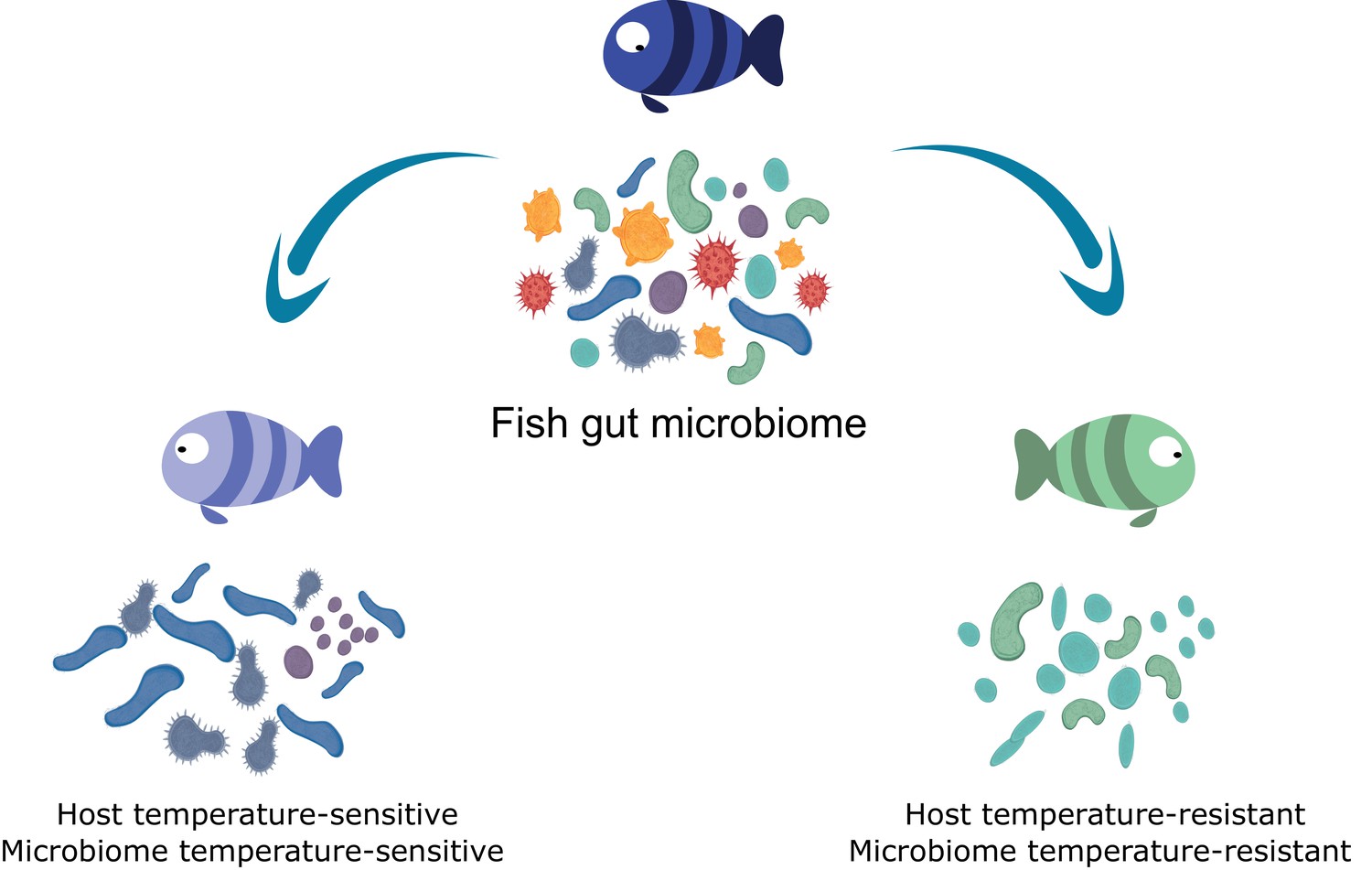
Our interests include community assembly dynamics of the microbiome in high temporal and genetic resolution. This enables us to identify the potential of alternative assemblages in gut communities, providing further information about basic ecological concepts. For example, a particular species may pre-condition future states of the community due to its ecological interactions, stimulating the growth of a specific subset of species and inhibiting others. We relate these variations in community composition to changes in ecological function, particularly the metabolic efficiency of the microbiome.
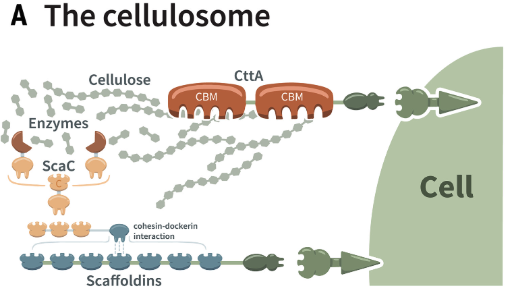
For as long as there has been life on land, Earth’s terrestrial food chain has been based on the microbial degradation of cellulose-based fiber of photosynthetic products. Although extensive research has been dedicated to the evolutionary origins and mechanisms of the photosynthetic processes of land plants that generate fiber, very little is known about the evolution and ecology of microbial fiber breakdown. We colboarte with Prof. William F. Martin, Prof. Ohad Medalia, Prof. Edward A. Bayer and Prof. Dörte Becher in a focused effort uniting cutting-edge research tools and expertise, by applying microbial ecology, extracellular proteomics, fiber-degrading enzymology, structural biology and molecular evolution, to understand the evolutionary history of fiber degradation and its interconnectivity with microbial ecology at a nanoscale resolution. Achieving our goals will enable us to gain answers and deep insights into how ecology and evolution of microbial fiber degradation are intertwined and reflected at the single-microbe and community context, from the biochemical, structural and physiological perspectives. Our study will thus provide a stepping-stone towards defining and better understanding the central role of fiber degradation in nature and will reveal the Earth history of natural fiber-degrading microbial ecosystems.
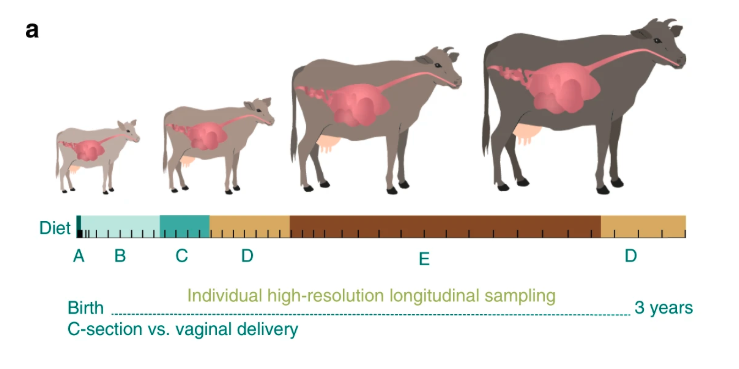
Ruminants host a complex relationship with their residing ruminal microbiota; a relationship which has evolved for millions of years and impacts our everyday lives regarding food availability, environmental issues and economic concerns. The microbiome resides in the upper digestive tract of the animal in a chambered compartment named the rumen and is mainly responsible for most of the food’s digestion and absorption. Therefore, understanding and characterizing this complex ecosystem is of major interest to us. Our results show that individual hosts vary greatly in the different pathways utilized by their microbiome for harvesting energy. Alternative pathways may conserve energy by maximizing energy production while suppressing methane emission- a quality termed ‘feed efficiency’. These pathways rely on species composition and the metabolic functions they provide. We bring together microbial genomics, anaerobic microbiology and microbial community ecology to learn how cooperative or antagonistic ecological interactions between the microbial species that reside in the rumen control the establishment of alternative microbiomes with different functional efficiencies.
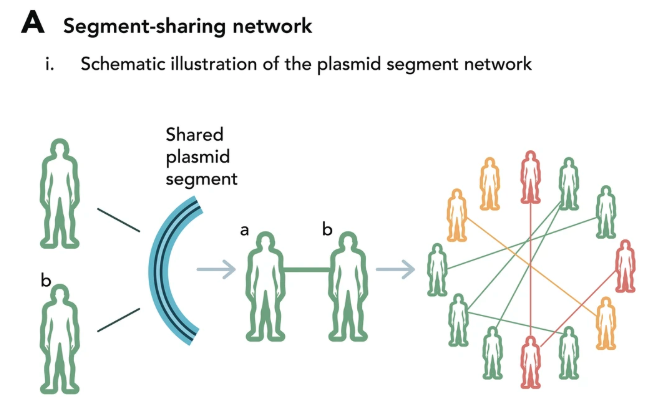
Gut microbial communities are extremely diverse, exceptionally dense and sustain an intricate relationship with their mammalian host. The immediate proximity and wide range of neighboring cells create a favorable environment for horizontal gene transfer (HGT). We focus on gene mobility as it rapidly changes genome plasticity and greatly influences microbial populations in nature and specifically, in gut environments. Plasmids are major mediators of HGT, contributing greatly to genome evolution. Virulence factors, as well as antibiotic-resistance genes, are frequently transferred on plasmids, enabling their rapid spread within and between bacterial populations. Our fascination with this topic, together with a lack of tools for its study, led us to develop a procedure of metagenomic plasmid isolation and characterization. This pioneering approach provides an overview of plasmids—their identity, traits and the phylogenetic diversity of their microbial hosts. Understanding the role of plasmids as carriers of genetic information is crucial to our understanding of microbial ecology and evolution.
Latest News
Seeing The Invisible Retread
This September, we held a three-day retreat at the Israel Institute for Advanced Studies (IIAS) in Jerusalem to refine our upcoming art exhibition with Maya Dvash, Shaul Cohen, Tamara Anna Efrat, and Ronit Milano.
During the retreat, we developed the various projects for the exhibition, focusing on multidisciplinarity with Professor Noam Mizrahi, Professor Itzik Hen & Dr. Yakov Merhishti.

The 2024 Bruno Prize has been granted to Prof. Itzhak Mizrahi
Itzik has been awarded the 2024 Bruno Prize for our research in microbial ecology. We are deeply honored and humbled by this recognition. Grateful to our amazing team and collaborators who made this possible.

Plasmids in the human gut reveal neutral dispersal and recombination that is overpowered by inflammatory diseases
We explored plasmid dispersal across 3467 human gut microbiome samples worldwide. Our findings show neutral gene flow, but under pressure, key genes breach barriers. Join us in uncovering insights at Nature Communication !
Novel Ruminococcal species in the human gut, proficient in cellulose degradation and finely tuned to our evolutionary journey.
We explored uncharted bacterial species in the human gut microbiota capable of degrading complex cellulosic polysaccharides, revealing their origins, adaptations to diverse host lifestyles and diets, and potential sources. Read more in Science.
Microbial war and peace hanging by a thread of a plasmid coded defense system
We discover that a plasmid coded defense system serves as a gateway for prosperous mutualistic interactions that otherwise could become waring and distractive ones.Check out our new paper at nature microbiology .
Behind-the-paper interview for FEMS
Check out this recent #BehindThePaper interview we conducted together with our collaborators from the Medalia Lab for #FEMSmicroBlog, discussing our recent work utilizing cryo-electron microscopy to gain new perspectives of microbial ecology and evolution
Phylogenetic diversity of core rumen microbiota described by cryo-ET
In our recent work published in microLIFE we used cryo-electron microscopy to capture the specific microscopic structure of microbes. We were able to classify microbes by their structure and create an atlas of rumen bacterial and archaeal species, combining both complementary fields of structural biology and microbial ecology.

Cell Voices
Check out the latest Cell Voices to read Itzik’s perspective on what collective features of microbial communities our lab is aiming to unravel.

New paper published in the ISME Journal
In this new paper published in the ISME Journal we find that the rumen metaproteome is remarkably plastic at the genome level & is predictive to microbiome & host feed efficiency states.
Nanoscale resolution of microbial fiber degradation in action
Looking through the microbial lens – Our recent work published in eLife, where we use single-cell structural biology to understand the ecology of cellulose degradation at scales in which microbial life unfolds

New collaborative paper with the Elia Lab
Check out our new paper published in the ISME Journal as part of a collaboration with the Elia Lab, where we study an Asgard archaeal protein family conserved across all domains of life.
New paper published in the ISME journal
Check out our new study published in the ISME journal, demonstrating how differences in gut microbial composition across lineages are translated into host-specific metabolomes, see the thread below from the main findings

Our new paper discussing the rumen microbiome is out in nature reviews microbiology!
Check out our recent review in Nature reviews. We describe the composition, ecology, and metabolism of the rumen microbiome and how to use it to balance food security and environmental impacts.
ERC Consolidator Grant
We got an ERC consolidator grant!!!!
Thank you, reviewers, and thank you, panel members, for enabling us to continue doing what we love!
Nice write up of on our work
Our new paper in Nature Communications in
Our new paper in Nature Communications. We found that the rumen microbiome has a long-term “memory” of early life events that could be used to manipulate and predict its assembly.

Science Advances
We might be able to modulate microbiomes using the cow genome. See our paper in Science Advances, in which we show that the cow genome is connected to its microbiome.![]()
Take a look at our post on the story behind our new paper in Nature communications at Nature blog
Where it all begins. A microbiome assembly story… Read our post on the story behind our new paper, published in Nature communications at Nature Microbiology Community
Paper in Nature Microbiology
Our new paper in Nature Microbiology gives a potential explanation of what core microbes are and how they maintain their stability in the gut.

elife
Check our paper in elife, where we found that genetic selection of the hosts also selects their microbiome and its response to environmental selection.
![]()
Publications

Who We Are?

Itzik Mizrahi
PI
Research interests: Microbial community assembly and evolution
Motto: “Do or do not, there is no try” -Master Yoda
Hobbies: Cooking

Sarah Morais
Staff Scientist
Research interests: Metabolic engineering of gut bacteria
Motto: Run your days and love what you do
Hobbies: Eating chocolate and mothering

Ori Furman
Staff Scientist
Research interests: Characterization of rumen bacteria
Motto: Big things come in small packages
Hobbies: Sailing and milking

Omar Tovar
Post-doc
Research interests: Microbial interactions
Motto: Everybody wants to go to heaven but nobody wants to die
Hobbies: Skateboarding, crossfit, hiking, reading

Omer Lavy
Post-doc
Research interests: Host-microbe interactions, bacterial interactions
Motto: Work hard and be nice to people
Hobbies: (who has time for hobbies??) Hiking,camping, running, cooking and five-needles knitting

Orna Schweltzer
Post-doc
Research interests: Isolation and Characterization of Rumen Microbiome
Motto: Feel the fear and do it anyway
Hobbies: Pilates, traveling, baking

Sarah Winkler
PhD Student
Research interests: Community assembly and synthetic rumen consortia
Motto: I have to be somewhere so it might as well be here
Hobbies: Cooking, yoga, travelling

Ido Grinshpan
PhD Student
Research interests: Engineering synthetic microbiomes
Motto: Believe no lies, pizza is the only superfood
Hobbies: Playing guitar, running, volleyball, powerlifting

Itai Amit
PhD Student
Research interests: Isolation and Characterization of Rumen Microbiome
Motto: Change through movement
Hobbies: Cooking, painting, pilates

Daniel Somekh
PhD Student
Research interests: Environmental factors and Microbe’s dynamics
Motto: “If you want a new idea, read an old book”
Hobbies: Lego, Puzzles and Hiking in the desert

Tomer Zur
PhD Student
Research interests: Disappearing Microbiome Hypothesis
Motto: Don’t Panic !
Hobbies: Collecting and hybridizing tropical plants, building bioactive terrariums and aquariums, and exploring traditional fermentation cuisine.
Mail: zurtom@post.bgu.ac.il

Jonathan Shelly
PhD Student
Research interests: Microbiome’s fundamentals and dynamics
Motto: “Never forget what you are. The rest of the world will not. Wear it like armour, and it can never be used to hurt you.” — Tyrion Lannister
Hobbies: Survive (And play the violin)

Tal Wilian
MSc Student
Research interests: Microbiome’s fundamentals and dynamics
Motto: Seize the day
Hobbies: Hiking, yoga and gardening
Mail: talwil@post.bgu.ac.il

Dvorah Babinski
Lab Technician
Research Interests: Sustainability of systems and climate change
Motto: Everything is for the good.
Hobbies: Spend time with family in nature.

Liel Gribun
Lab Technician
Research Interests: Plasmids and microbial communities
Motto: You don’t have to feel ready to begin
Hobbies: Collecting and growing unique plants, cooking and baking, aerial acrobatics.

Malka Kichikova
Lab Technician
Research Interests: Isolation and Characterization of Rumen Microbiome
Motto: “A day without learning is a day wasted.” – Albert Einstein
Hobbies: Baking, Fire spinning, and Yoga

Tanya Karakis
Administrative Manager
Motto: “Never give up on your dreams, because that’s where the magic begins.” (Willy Wonka)
Hobbies: 🎶 Music – I love to sing, dance, and listen to songs. ⛸️ Ice skating, spending time with my kids (being a mom is definitely a favorite hobby! 😍), and also cooking delicious things in the kitchen.

Zoe Coleman Becker
BSc Student
Motto: Live life to its fullest
Hobbies: Yoga, traveling and reading

Maya Hascalovich
BSc Student
Motto: “Everything goes wrong for a reason”
Hobbies: Music and Trips around the world
Mail: Mayahasc@post.bgu.ac.il

Tanya Geystrin
BSc Student
Motto: Where words fail, music speaks
Hobbies: Climbing, painting, riding books and listening to music
Lab Life
Alumni
Dr. Tamar Zehavi
Currently: Company scientist at “1e Therapeutics”
Motto: Short people are just the same as you and I
Dr. Elie Jami
Dr. Fotini Kokou
Currently: Assistant professor Whchinigen university Netherlands
Motto: You can always swim your way out of it
Inbal Ashkenazi
Currently: Scientist at “Evogene”
Motto: Kindness is a virtue
Michael Mazor
Currently: Food Technologist in R&D team
Motto: Keep your eyes on the stars and your feet on the ground
Eran Gavrieli
Motto: Steady yourself by supporting others
Ido Lybovits
Currently: Research Assistant at biomica ltd.
Motto: Take the risk or lose the chance
Gil Sorek
Motto: Do what you love and love what you do
Sapir Nadler
Currently: Medical Doctor student in TAU
Motto: Life’s too mysterious to take too serious
Daphne Perlman
Currently: Scientific illustrator and graphic designer
Motto: Those who dare to fail miserably can achieve greatly
Gal Revivo
Motto: “Every day it gets a little easier… But you gotta do it every day — that’s the hard part. But it does get easier”.
Yoav Shaani
Eran Shriker
Currently: CTO & Co-Founder – BioTech Elements
Motto: “Ask, believe, receive” -Rhonda Byrne “The Secret”
Dr. David Bogumil
Currently: Bioinformatics Analyst at BiomX Ltd
Motto: Beer. Now there’s a temporary solution.
Dr. Sheerli Kruger Ben Shabat
Currently: Head of computational human microbiome research at Biomica Ltd
Motto: The only way is up
Dr. Nir Friedman
Dr. Thomer Durman
Currently: Profesor university of Estadual de Maringá Brazil
Motto: I did not say it would be easy, I said it would worth it
Stav Eyal
Currently: Researcher at BiomX Ltd
Motto: Just flow
Dr. Maraike Probst
Research interests: Rumen plasmids
Motto: Phenomenal cosmic powers. Itty-bitty living space
Hobbies: Moving fast
Dr. Sasha Kugel
Research interests: Genome Assembly
Motto: Stand back – I’m gonna try science!
Hobbies: Drowning (water, data, ice-cream, you name it…)
Naama Shterzer
Research interests: Mobilizable plasmids
Motto: Procrastinate now, don’t put it off
Hobbies: Playing squash
Dr. Aya Brown Kav
Research interests: Characterization of rumen plasmids
Motto: Get to the point
Hobbies: Kite surfing
Goor Sasson
Research interests: Rumen metagenomics
Motto: There must be an easier and smarter way to do it
Hobbies: In my spare time I like to practice modern dance
Adi Israel
Research interests: Characterization of rumen methanogens
Motto: There is order in everything
Hobbies: Smiling
Eitai Landou
Research interests: Metabolic engineering
Motto: There is order in chaos
Hobbies: Collecting reptiles
Nili Dagan
Currently: Administrative assistant at BGU
Motto: Always see the full half of the glass
Chagai Davidovich
Motto: You cannot discover new oceans unless you have the courage to lose sight of the shore
Hobbies: Traveling, basketball, playing guitar

Courses

Computational Microbial Ecology

Microbial Ecology
Contact Us
Address
Ben-Gurion University of the Negev, Faculty of Natural Sciences, Life Sciences, building 41, room 228
Phone
08-6479836/7/8
Fax
08-6479839
Mail
imizrahi@bgu.ac.il