2025
Julian Herz, Carina Weigel, Leonie Scheder, Raz Zarivach, Itay Aglov, Yonatan Chemla, Felix Popp, Cornelius Riese, Mohammad A Charsooghi, Lital Alfonta, Michael M Meijler, Dirk Schüler, Damien Faivre, Daniel Pfeiffer
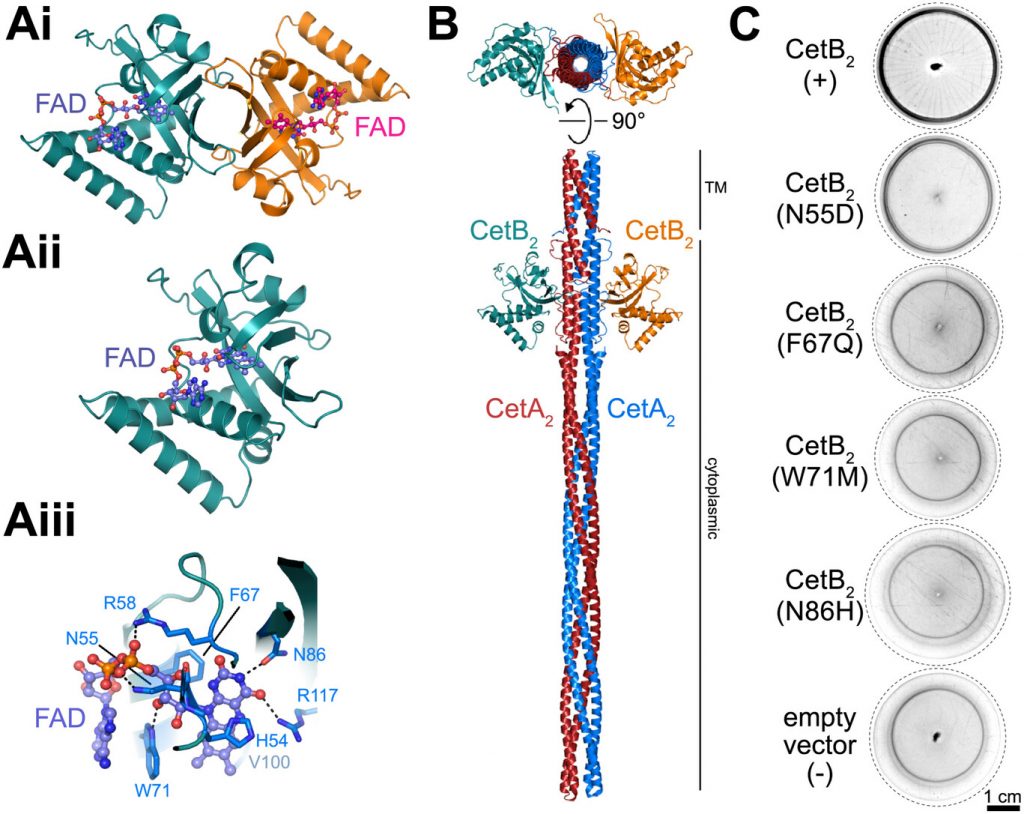
A Two-Protein Chemoreceptor Complex Regulates Oxygen Thresholds in Bacterial Magneto-Aerotaxis
2024
Moran Elias-Mordechai, May Morhaim, Maya Georgia Pelah, Irina Rostovsky, May
Nogaoker, Jürgen Jopp, Raz Zarivach, Neta Sal-Man, Ronen Berkovich

2023
Tomer Eli Ben Yosef, Raz Zarivach, Arie Moran
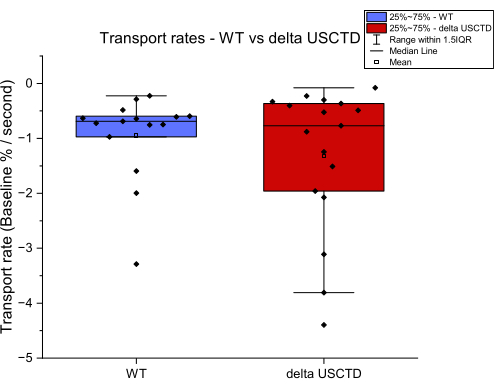
Characterizing Mammalian Zinc Transporters Using an In Vitro Zinc Transport Assay
10.3791/65217
Pierre Majdalani *,Uri Yoel*, Tayseer Nasasra, Merav Fraenkel, Alon Haim, Neta Loewenthal, Raz Zarivach, Eli Hershkovitz, and Ruti Parvari
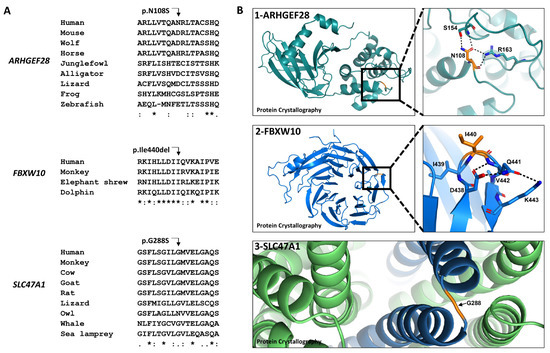
Novel Susceptibility Genes Drive Familial Non-Medullary Thyroid Cancer in a Large Consanguineous Kindred
https://doi.org/10.3390/ijms24098233
Yuval Yogev , Zamir Shorer, Arie Koifman, Ohad Wormser, Max Drabkin , Daniel Halperin , Vadim Dolgin, Regina Proskorovski-Ohayon, Noam Hadar , Geula Davidov , Hila Nudelman, Raz Zarivach , Ilan Shelef, Yonatan Perez , and Ohad S. Birk
Limb girdle muscular disease caused by HMGCR mutation and statin myopathy treatable with mevalonolactone
https://doi.org/10.1073/pnas.2217831120

Pierre Majdalani *, Aviva Levitas *, Hanna Krymko, Leonel Slanovic, Alex Braiman, Uzi Hadad, Salam Dabsan, Amir Horev, Raz Zarivach, Ruti Parvari
A Missense Variation in PHACTR2 Associates with Impaired Actin Dynamics, Dilated Cardiomyopathy, and Left Ventricular Non-Compaction in Humans
https://doi.org/10.3390/ijms24021388
2022
Dikla Nachmias, Nataly Melnikov, Alvah Zorea, Yasmin De-picchoto, Raz Zarivach, Itzhak Mizrahi, Natalie Elia
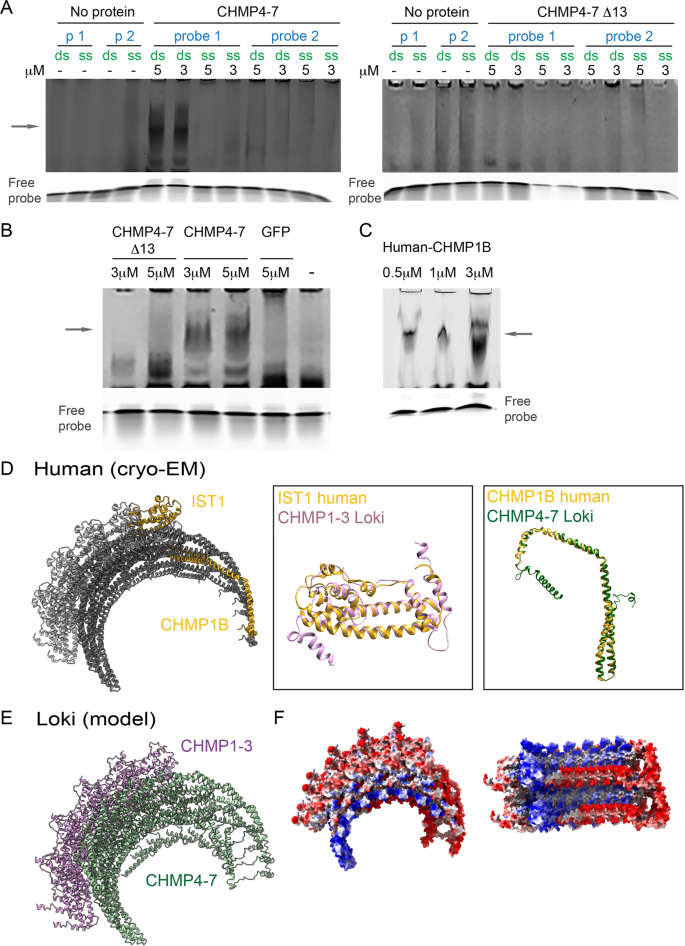
Asgard ESCRT-III and VPS4 reveal evolutionary conserved chromatin binding properties of the ESCRT machinery
https://doi.org/10.1101/2021.11.29.470303
Nila Nandha Kadamannil , Jung-Moo Heo , Daewoong Jang , Ran Zalk , Sofiya Kolusheva , Raz Zarivach , Gabriel A Frank , Jong-Man Kim , Raz Jelinek
High-Resolution Cryo-Electron Microscopy Reveals the Unique Striated Hollow Structure of Photocatalytic Macrocyclic Polydiacetylene Nanotubes
https://doi.org/10.1021/jacs.2c06710
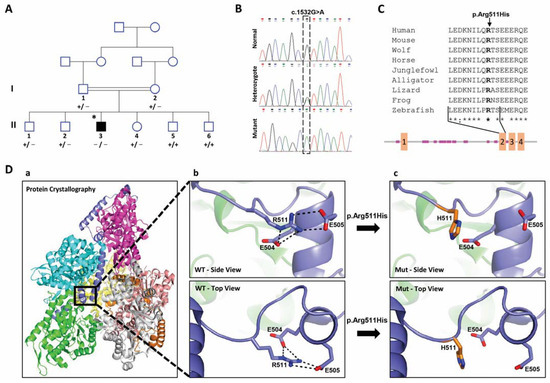
PSMC1 variant causes a novel neurological syndrome
Ramesh Kumar Krishnan , Naomi Halachmi , Raju Baskar , Anna Bakhrat , Raz Zarivach , Adi Salzberg , Uri Abdu
Revisiting the Role of ß-Tubulin in Drosophila Development: β-tubulin60D is not an Essential Gene, and its Novel Pin1 Allele has a Tissue-Specific Dominant-Negative Impact
https://doi.org/10.3389/fcell.2021.787976
Shannon L Kelleher, Samina Alam, Olivia C Rivera, Shiran Barber-Zucker, Raz Zarivach, Takumi Wagatsuma, Taiho Kambe, David I Soybel, Justin Wright &Regina Lamendella
Loss-of-function SLC30A2 mutants are associated with gut dysbiosis and alterations in intestinal gene expression in preterm infants
https://doi.org/10.1080/19490976.2021.2014739
2021
Anna Pohl , Sarah A E Young , Tara C Schmitz , Daniel Farhadi , Raz Zarivach , Damien Faivre , Kerstin G Blank
Magnetite-binding proteins from the magnetotactic bacterium Desulfamplus magnetovallimortis BW-1Current view of iron biomineralization in magnetotactic bacteria
PMID: 34860229 DOI: 10.1039/d1nr04870h
Shirel Ben-Shimon *, Daniel Stein* , Raz Zarivach
Current view of iron biomineralization in magnetotactic bacteria
RSC Chem Biol. 2021. PMID: 34458794

Simon-Baram H, Kleiner D, Shmulevich F, Zarivach R, Zalk R, Tang H, Ding F, Bershtein S.
SAMase of Bacteriophage T3 Inactivates Escherichia coli’s Methionine S-Adenosyltransferase by Forming Heteropolymers.
mBio. 2021 Aug3:e0124221

Pandey H, Singh SK, Sadan M, Popov M, Singh M, Davidov G, Inagaki S, Al-Bassam J, Zarivach R, Rosenfeld SS, Gheber L.
Flexible microtubule anchoring modulates the bi-directional motility of the kinesin-5 Cin8.
Cell Mol Life Sci. 2021 Jul 17.
Juan Camilo Moreno, Bruno E Rojas, Rubén Vicente, Michal Gorka, Timon Matz, Monika Chodasiewicz, Juan S Peralta-Ariza, Youjun Zhang, Saleh Alseekh, Dorothee Childs, Marcin Luzarowski, Zoran Nikoloski, Raz Zarivach, Dirk Walther, Matías D Hartman, Carlos M Figueroa, Alberto A Iglesias, Alisdair R Fernie, Aleksandra Skirycz
Tyr‐Asp inhibition of glyceraldehyde 3‐phosphate dehydrogenase affects plant redox metabolism.
The EMBO Journal (2021): e106800.
Baron N, Tupperwar N, Dahan I, Hadad U, Davidov G, Zarivach R, Shapira M.
Distinct features of the Leishmania cap-binding protein LeishIF4E2 revealed by CRISPR-Cas9 mediated hemizygous deletion.
PLoS Negl Trop Dis. 2021 Mar 24;15(3):e0008352.

aingelernt Y, Hershkovitz E, Abu-Libdeh B, Abedrabbo A, Abu-Rmaileh Amro S, Zarivach R, Zangen D, Lavi E, Haim A, Parvari R, Abu-Libdeh A.
Aldosterone synthase (CYP11B2) deficiency among Palestinian infants: Three novel variants and genetic heterogeneity. Am J Med Genet A. 2021 Jan 13. doi: 10.1002/ajmg.a.62056. Epub ahead of print. PMID:
Davidov G, Abelya G, Zalk R, Izbicki B, Shaibi S, Spektor L, Shagidov D, Meyron-Holtz EG, Zarivach R, Frank GA.
Folding of an Intrinsically Disordered Iron-Binding Peptide in Response to Sedimentation Revealed by Cryo-EM. J Am Chem Soc. 2020 Nov 9. doi: 10.1021/jacs.0c07565. Epub ahead of print. PMID: 33166133.
2020
Barber-Zucker S, Hall J, Froes A, Kolusheva S, MacMillan F, Zarivach R.
The cation diffusion facilitator protein MamM’s cytoplasmic domain exhibits metal-type dependent binding modes and discriminates against Mn2. J Biol Chem. 2020 Sep 23:jbc.RA120.014145. doi: 10.1074/jbc.RA120.014145. Epub ahead of print. PMID: 32967967.

Barber-Zucker S, Shahar A, Kolusheva S, Zarivach R.
The metal binding site composition of the cation diffusion facilitator protein MamM cytoplasmic domain impacts its metal responsivity. Sci Rep. 2020 Aug 20;10(1):14022. doi: 10.1038/s41598-020-71036-4. PMID: 32820200; PMCID: PMC7441159.

Pizarro L, Leibman-Markus M, Gupta R, Kovetz N, Shtein I, Bar E, Davidovich-Rikanati R, Zarivach R, Lewinsohn E, Avni A, Bar M.
A gain of function mutation in SlNRC4a enhances basal immunity resulting in broad-spectrum disease resistance. Commun Biol. 2020 Jul 30;3(1):404. doi: 10.1038/s42003-020-01130-w. PMID: 32732974; PMCID: PMC7393091.

Rivera OC, Geddes DT, Barber-Zucker S, Zarivach R, Gagnon A, Soybel DI, Kelleher SL.
A common genetic variant in zinc transporter ZnT2 (Thr288Ser) is present in women with low milk volume and alters lysosome function and cell energetics. Am J Physiol Cell Physiol. 2020 Jun 1;318(6):C1166-C1177. doi: 10.1152/ajpcell.00383.2019. Epub 2020 Apr 22. PMID:32320289.

Barber-Zucker S, Shahar A, Kolusheva S, Zarivach R.
The metal binding site composition of the cation diffusion facilitator protein MamM cytoplasmic domain impacts its metal responsivity. Sci Rep. 2020;10(1):14022. Published 2020 Aug 20. doi:10.1038/s41598-020-71036-4

Keren-Khadmy N, Zeytuni N, Kutnowski N, Perriere G, Monteil C, Zarivach R.
From conservation to structure, studies of magnetosome associated
cation diffusion facilitators (CDF) proteins in Proteobacteria. PLoS One. 2020;15(4):e0231839. Published 2020 Apr 20. doi:10.1371/journal.pone.0231839.
Chen YC, Chen SP, Li JY, Chen PC, Lee YZ, Li KM, Zarivach R, Sun YJ, Sue SC. Integrative model to coordinate the oligomerization and aggregation mechanisms of CCL5. J Mol Biol. pii: S0022-2836(20)30038-3.
2019
Tseytin I, Mitrovic B, David N, Langenfeld K, Zarivach R, Diepold A, Sal-Man N. The Role of the Small Export Apparatus Protein, SctS, in the Activity of the Type III Secretion System. Front Microbiol. 10:2551.
Drabkin M, Yogev Y, Zeller L, Zarivach R, Zalk R, Halperin D, Wormser O, Gurevich E, Landau D, Kadir R, Perez Y, Birk OS.
Hyperuricemia and gout caused by missense mutation in d-lactate dehydrogenase. J Clin Invest. 2019 Dec 2;129(12):5163-5168. doi: 10.1172/JCI129057. PMID: 31638601; PMCID: PMC6877321.

Kleiner D, Shmulevich F, Zarivach R, Shahar A, Sharon M, Ben-Nissan G, Bershtein S.
The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase. J Mol Biol. 2019 Dec 6;431(24):4796-4816. doi: 10.1016/j.jmb.2019.09.003. Epub 2019 Sep 12. PMID: 31520601.


Unexpected implications of STAT3 acetylation revealed by genetic encoding of acetyl-lysine. Biochim Biophys Acta Gen Subj. 2019 Sep;1863(9):1343-1350. doi: 10.1016/j.bbagen.2019.05.019. Epub 2019 Jun 3. PMID: 31170499; PMCID: PMC6625855.

Taparia Y, Zarka A, Leu S, Zarivach R, Boussiba S, Khozin-Goldberg I.
A novel endogenous selection marker for the diatom Phaeodactylum tricornutum based on a unique mutation in phytoene desaturase 1. Sci Rep. 2019 Jun 3;9(1):8217. doi: 10.1038/s41598-019-44710-5. PMID: 31160749; PMCID: PMC6546710.

Kleiner D, Shmulevich F, Zarivach R, Shahar A, Sharon M, Ben-Nissan G, Bershtein S. The inter-dimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase. Now in bioRxiv (2019). doi: https://doi.org/10.1101/669069.
Belo Y, Mielko Z, Nudelman H, Afek A, Ben-David O, Shahar A, Zarivach R, Gordan R, Arbely E. Unexpected implications of STAT3 acetylation revealed by genetic encoding of acetyl-lysine. Biochim Biophys Acta Gen Subj (2019). doi: 10.1016/j.bbagen.2019.05.019.

Baskar R, Bahkrat A, Otani T, Wada H, Davidov G, Pandey H, Gheber L, Zarivach R, Hayashi S, Abdu U. The plus-tip tracking and microtubule stabilizing activities of Javelin-like regulate microtubule organization and cell polarity. FEBS J (2019). doi: 10.1111/febs.14944.

Levy M, Elkoshi N, Barber-Zucker S, Hoch E, Zarivach R, Hershfinkel M, Sekler I. Zinc transporter 10 (ZnT10)-dependent extrusion of cellular Mn2+ is driven by an active Ca2+-coupled exchange. J Biol Chem (2019). doi: 10.1074/jbc.RA118.006816.

Aspit L, Levitas A, Etzion S, Krymko H, Slanovic L, Zarivach R, Etzion Y, Parvari R. CAP2 mutation leads to impaired actin dynamics and associates with supraventricular tachycardia and dilated cardiomyopathy. J Med Genet (2018). doi: 10.1136/jmedgenet-2018-105498.
2018
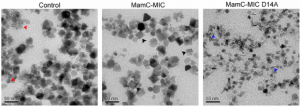
Nudelman H, Lee YZ, Hung YL, Kolusheva S, Upcher A, Chen YC, Chen JY, Sue SC, Zarivach R. Understanding the Biomineralization Role of Magnetite-Interacting Components (MICs) From Magnetotactic Bacteria. Front Microbiol (2018) 9:2480. doi: 10.3389/fmicb.2018.02480.
Gitz JC, Sadot N, Zaccai M, Zarivach R. A Calorimetric Method for Measuring Iron Content in Plants. J Vis Exp (2018) 7:(139). doi: 10.3791/57408.

Kutnowski N, Shmuely H, Dahan I, Shmulevich F, Davidov G, Shahar A, Eichler J, Zarivach R, Shaanan B. The 3-D structure of VNG0258H/RosR – a haloarchaeal DNA-binding protein in its ionic shell. J Struct Biol (2018) In Press. doi.org/10.1016/j.jsb.2018.08.008.
Gertman O, Omer D, Hendler A, Stein D, Onn L, Khukhin Y, Portillo M, Zarivach R, Cohen HY, Toiber D, Aharoni A. Directed evolution of SIRT6 for improved deacylation and glucose homeostasis maintenance. Sci Rep (2018) 8(1):3538.doi:10.1038/s41598-018-21887-9.
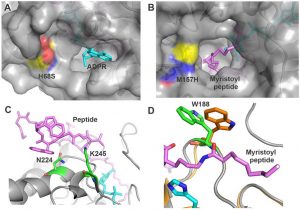
Abehsera S, Zaccai S, Mittelman B, Glazer L, Weil S, Khalaila I, Davidov G, Bitton R, Zarivach R, Li S, Li F, Xiang J, Manor R, Aflalo ED, Sagi A. CPAP3 proteins in the mineralized cuticle of a decapod crustacean. Sci Rep (2018) 8(1):2430. doi: 10.1038/s41598-018-20835-x.
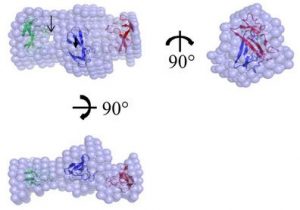
Chen YC, Li KM, Zarivach R, Sun YJ, Sue SC. Human CCL5 trimer: expression, purification and initial crystallographic studies. Acta Cryst F (2018) 74(Pt 2):82-85. doi: 10.1107/S2053230X17018544.

Nudelman H, Perez Gonzalez T, Kolushiva S, Widdrat M, Reichel V, Peigneux A, Davidov G, Bitton R, Faivre D, Jimenez-Lopez C, Zarivach R. The importance of the helical structure of a MamC-derived magnetite-interacting peptide for its function in magnetite formation. Acta Cryst D (2018) 74(Pt 1):10-20. doi: 10.1107/S2059798317017491.
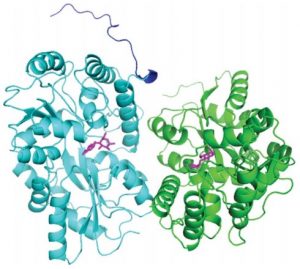
Uebe R, Keren-Khadmy N, Zeytuni N, Katzmann E, Navon Y, Davidov G, Bitton R, Plitzko JM, Schüler D, Zarivach R.
The dual role of MamB in magnetosome membrane assembly and magnetite biomineralization. Mol Microbiol. 2018 Feb;107(4):542-557. doi: 10.1111/mmi.13899. Epub 2018 Jan 9. PMID: 29243866.
Uebe R*, Keren-Khadmy N*, Zeytuni N, Katzmann E, Navon Y, Davidov G, Bitton R, Plitzko JM, Schüler D, Zarivach R. The dual role of MamB in magnetosome membrane assembly and magnetite biomineralization. Mol Microbiol (2017) 107(4):542-557. doi: 10.1111/mmi.13899.
2017
Barber-Zucker S, Shaanan B, Zarivach R. Transition metal binding selectivity in proteins and its correlation with the phylogenomic classification of the cation diffusion facilitator protein family. Sci Rep (2017) 7(1):16381. doi: 10.1038/s41598-017-16777-5.
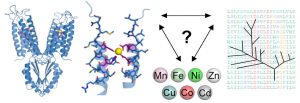
Raschdorf O, Bonn F, Zeytuni N, Zarivach R, Becher D, Schüler D. A quantitative assessment of the membrane-integral sub-proteome of a bacterial magnetic organelle. J Proteomics (2017) pii: S1874-3919(17)30360-3. doi: 10.1016/j.jprot.2017.10.007.
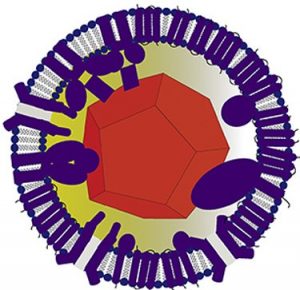
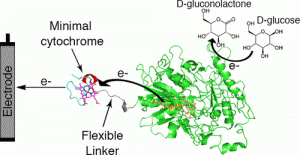
Algov I, Grushka J, Zarivach R, Alfonta L. Highly efficient flavin-adenine dinucleotide glucose dehydrogenase fused to a minimal cytochrome c domain. J Am Chem Soc (2017) Epub ahead of print. doi: 10.1021/jacs.7b07011.
Perez Y, Shorer Z, Liani-Leibson K, Chabosseau P, Kadir R, Volodarsky M, Halperin D, Barber-Zucker S, Shalev H, Schreiber R, Gradstein L, Gurevich E, Zarivach R, Rutter GA, Landau D, Birk OS. SLC30A9 mutation affecting intracellular zinc homeostasis causes a novel cerebro-renal syndrome. Brain (2017) 140(4):928-939. doi: 10.1093/brain/awx013.

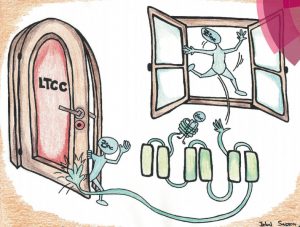
Shusterman E, Beharier O, Levy S, Zarivach R, Etzion Y, Campbell CR, Lee IH, Dinudom A, Cook DI, Peretz A, Katz A, Gitler D and Moran A. Zinc transport and the inhibition of the L-type calcium channel are two separable functions of ZnT-1. Metallomics (2017) 9(3):228-238. doi: 10.1039/C6MT00296J.
2016
Barber-Zucker S, Zarivach R. A Look into the Biochemistry of Magnetosome Biosynthesis in Magnetotactic Bacteria. ACS Chem Biol (2016) 12(1):13-22. doi: 10.1021/acschembio.6b01000.
Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, Blumberg A, Schlesinger O, Bieri P, Greber B, Ban N, Zarivach R, Alfonta L, Pilpel Y, Suzuki T, Mishmar D. Mitochondrial 16S rRNA Is Methylated by tRNA Methyltransferase TRMT61B in All Vertebrates. PLoS Biol (2016) 14(9):e1002557. doi: 10.1371/journal.pbio.1002557.

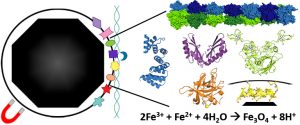
Barber-Zucker S, Uebe R, Davidov G, Navon Y, Sherf D, Chill JH, Kass I, Bitton R, Schüler D, Zarivach R. Disease-Homologous Mutation in the Cation Diffusion Facilitator Protein MamM Causes Single-Domain Structural Loss and Signifies Its Importance. Sci Rep (2016) 6:31933. doi: 10.1038/srep31933.

Regev O, Korman M, Hecht N, Roth Z, Forer N, Zarivach R, Gur E. An extended loop of the Pup ligase, PafA, mediates interaction with protein targets. J Mol Biol (2016) pii: S0022-2836(16)30288-1. doi: 10.1016/j.jmb.2016.07.021.

Auerbach-Nevo T, Baram D, Bashan A, Belousoff M, Breiner E, Davidovich C, Cimicata G, Eyal Z, Halfon Y, Krupkin M, Matzov D, Metz M, Rufayda M, Peretz M, Pick O, Pyetan E, Rozenberg H, Shalev-Benami M, Wekselman I, Zarivach R, Zimmerman E, Assis N, Bloch J, Israeli H, Kalaora R, Lim L, Sade-Falk O, Shapira T, Taha-Salaime L, Tang H, Yonath A. Ribosomal Antibiotics: Contemporary Challenges. Antibiotics (2016) 5(3):24 doi:10.3390/antibiotics5030024.
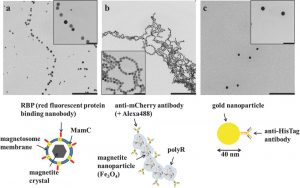
Jehle F, Valverde-Tercedor C, Reichel V, Carillo MA, Bennet M, Günther E, Wirth R, Mickoleit F, Zarivach R,
Schüler D, Blank KS, Faivre D. Genetically Engineered Organization: Protein Template, Biological Recognition Sites, and Nanoparticles. Adv Mater Interface (2016) doi: 10.1002/admi.201600285.
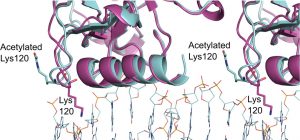
Vainer R, Cohen S, Shahar A, Zarivach R, Arbely E. Structural basis for p53 Lys120-acetylation dependent DNA-binding mode. J Mol Biol (2016) pii: S0022-2836(16)30217-0. doi: 10.1016/j.jmb.2016.06.009.
Radoul M, Lewin L, Cohen B, Oren R, Popov S, Davidov G, Vandsburger MH, Harmelin A, Bitton R, Greneche JM, Neeman M, Zarivach R. Genetic manipulation of iron biomineralization enhances MR relaxivity in a ferritin-M6A chimeric complex. Sci Rep (2016) 6:26550. doi: 10.1038/srep26550.

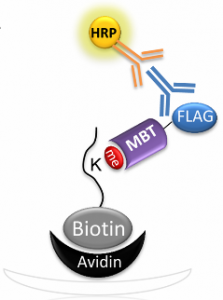
Cohen DO, Duchin S, Feldman M, Zarivach R, Aharoni A, Levy D. Engineering of Methylation State Specific 3xMBT Domain Using ELISA Screening. PLoS One (2016) 11(4):e0154207.
Nudelman H, Tercedor CV, Kolusheva S, Gonzalez TP, Widdrat M, Grimberg N, Levi H, Nelkenbaum O, Davidov G, Faivre D, Jimenez-Lopez C, Zarivach R. Structure-function studies of the magnetite-biomineralizing magnetosome-associated protein MamC. J Struct Biol (2016) 194(3):244-52. doi: 10.1016/j.jsb.2016.03.001.
Barber-Zucker S*, Keren-Khadmy N*, Zarivach R. From invagination to navigation: The story of magnetosome-associated proteins in magnetotactic bacteria. Protein Sci (2016) 25(2):338-51. doi: 10.1002/pro.2827].
2015
Bar-Yaacov D, Hadjivasiliou Z, Levin L, Barshad G, Zarivach R, Bouskila A, Mishmar D. Mitochondrial involvement in vertebrate speciation? The case of mito-nuclear genetic divergence in chameleons. Genome Biol Evol (2015) 7(12):3322-36. doi: 10.1093/gbe/evv226.
Zeytuni N*, Cronin S*, Lefèvre CT, Arnoux P, Baran D, Shtein Z, Davidov G, Zarivach R. MamA as a Model Protein for Structure-Based Insight into the Evolutionary Origins of Magnetotactic Bacteria. PLoS One (2015) 10(6):e0130394. doi: 10.1371/journal.pone.0130394. eCollection 2015.
Davidov G, Müller FD, Baumgartner J, Bitton R, Faivre D, Schüler D, Zarivach R.Crystal structure of the magnetobacterial protein MtxA C-terminal domain reveals a new sequence-structure relationship. Front Mol Biosci (2015) 2:25. doi: 10.3389/fmolb.2015.00025. eCollection 2015.
2014
Shusterman E, Beharier O, Shiri L, Zarivach R, Etzion Y, Campbell CR, Lee IH, Okabayashi K, Dinudom A, Cook DI, Katz A and Moran A. ZnT-1 extrudes zinc from mammalian cells functioning as a Zn2+/H+ exchanger. Metallomics (2014) 6(9):1656-63. doi: 10.1039/c4mt00108g.
Zeytuni N*, Uebe R*, Maes M, Davidov G, Baram M, Raschdorf O, Friedler A, Miller Y, Schüler D, Zarivach R. Bacterial Magnetosome Biomineralization – A Novel Platform to Study Molecular Mechanisms of Human CDF-Related Type-II Diabetes. PLoS One (2014) 9(5):e97154. doi:10.1371/journal.pone.0097154. eCollection 2014.
Zeytuni N*, Uebe R*, Maes M, Davidov G, Baram M, Raschdorf O, Nadav-Tsubery M, Kolusheva S, Bitton R, Goobes G, Friedler A, Miller Y, Schüler D, Zarivach R. Cation diffusion facilitators transport initiation and regulation is mediated by cation induced conformational changes of the cytoplasmic domain. PLoS One (2014) 9(3):e92141. doi: 10.1371/journal.pone.0092141. eCollection 2014.
Nudelman H, Zarivach R. Structure prediction of magnetosome-associated proteins. Front Microbiol (2014) 5:9. doi: 10.1002/chem.201300389.
Guttman C, Davidov G, Yahalom A, Shaked H, Kolusheva S, Bitton R, Barber-Zucker S, Chill JH, Zarivach R. BtcA, A Class IA Type III Chaperone, Interacts with the BteA N-Terminal Domain through a Globular/Non-Globular Mechanism. PLoS One (2014) 8(12):e81557. doi: 10.1371/journal.pone.0081557. eCollection 2013.
Müller FD, Raschdorf O, Nudelman H, Messerer M, Katzmann E, Plitzko JM, Zarivach R, Schüler D. The FtsZ-like protein FtsZm of Magnetospirillum gryphiswaldense likely interacts with its generic homolog and is required for biomineralization under nitrate deprivation. J Bacteriol (2014) 196(3):650-9. doi: 10.1128/JB.00804-13.
2013
.Kshirsagar UA, Parnes R, Goldshtein H, Ofir R, Zarivach R, Pappo D. Aerobic Iron-Based Cross-Dehydrogenative Coupling Enables Efficient Diversity-Oriented Synthesis of Coumestrol-Based Selective Estrogen Receptor Modulators. Chemistry (2013) 19(40):13575-83. doi: 10.1002/chem.201300389.
Dadon Z, Samiappan M, Shahar A, Zarivach R, Ashkenasy G. A High-Resolution Structure that Provides Insight into Coiled-Coil Thiodepsipeptide Dynamic Chemistry. Angewandte Chemie International Edition (2013) 52(38)9944-7. doi: 10.1002/anie.201303900. [Epub ahead of print].
Bar-Yaacov D, Avital G, Levin L, Richards A, Hachen N, Rebolledo Jaramillo B, Nekrutenko A, Zarivach R, Mishmar D. RNA-DNA differences in human mitochondria restore ancestral form of 16S ribosomal RNA. Genome Research (2013) 23(11)1789-96. doi:10.1101/gr.161265.113.
Guttman C, Davidov G, Shaked H, Kolusheva S, Bitton R, Ganguly A, Miller JF, Chill JH, Zarivach R. Characterization of the N-Terminal Domain of BteA: A Bordetella Type III Secreted Cytotoxic Effector. PLoS ONE (2013) 8(1): e55650. doi:10.1371/journal.pone.0055650.
2012
Zeytuni N, Baran D, Davidov G, Zarivach R. Inter-phylum structural conservation of the magnetosome-associated TPR-containing protein, MamA. J Struct Biol (2012) 180(3):479-87. doi: 10.1016/j.jsb.2012.08.001.
Zeytuni N, Offer T, Davidov G, Zarivach R. Crystallization and preliminary crystallographic analysis of the C-terminal domain of MamM, a magnetosome-associated protein from Magnetospirillum gryphiswaldense MSR-1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Aug 1;68(Pt 8):927-30. Epub 2012 Jul 31.
Regev T, Myers N, Zarivach R, Fishov I. Association of the Chromosome Replication Initiator DnaA with the Escherichia coli Inner Membrane In Vivo: Quantity and Mode of Binding. PLoS One. 2012;7(5):e36441. Epub 2012 May 4.
Zeytuni N, Zarivach R. The TPR Motif as a Protein Interaction Module – A Discussion of Structure and Function, Protein Interactions, Jianfeng Cai & Rongsheng E. Wang (Ed.), 2012, ISBN: 978-953-51-0244-1, InTech.
Zeytuni N, Zarivach R. Structural and Functional Discussion of the Tetra-Trico-Peptide Repeat, a Protein Interaction Module. Structure, Volume 20, Issue 3, 397-405, 7 March 2012.
2011
Berger I*, Guttman C*, Amar D, Zarivach R, Aharoni A. The Molecular Basis for the Broad Substrate Specificity of Human Sulfotransferase 1A1. 2011 PLoS One. 6(11): e26794. doi:10.1371/journal.pone.0026794
Uebe R, Junge K, Henn V, Poxleitner G, Katzmann E, Plitzko JM, Zarivach R, Kasama T, Wanner G, Posfai M, Bottger L, Matzanke B, Schuler D. The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions, and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol Microbiol. 2011 Oct 18. doi: 10.1111/j.1365-2958.2011.07863.x.
Zeytuni N, Ozyamak E, Harush KB, Davidov G, Levin M, Gat Y, Moyal T, Brik A, Komeili A, Zarivach R.. Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly. Proc Natl Acad Sci USA. 2011 Jul 22.
2010
Baruch K, Gur-Arie L, Nadler C, Koby S, Yerushalmi G, Ben-Neriah Y, Yogev O, Shaulian E, Guttman C, Zarivach R, Rosenshine I. Metalloprotease type III effectors that specifically cleave JNK and NF-κB. EMBO J. 2010 30(1),221-231.
Zeytuni N, Zarivach R. Crystallization and preliminary crystallographic analysis of the Magnetospirillum magneticum AMB-1 and M. gryphiswaldense MSR-1 magnetosome-associated proteins MamA. Acta Cryst. (2010). F66, 824-827.
Zeytuni N, Zarivach R. Purification of the M. magneticum Strain AMB-1 Magnetosome Associated Protein MamAΔ41. . J Vis Exp. 37 (2010)
2009
Cheung TYS, Fairchild MJ, Zarivach R, Tanentzapf G, Van Petegem F. Crystal Structure of the Talin Integrin Binding Domain. J Mol. Bio. 387(4):787-793 (2009).
2008
Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NC. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature. 453, 124-127 (2008).
2007
Zarivach R, Vuckovic M, Deng W, Finlay BB, Strynadka NCJ. Structural analysis of a prototypical ATPase from the type III secretion system. Nat Struct Mol Biol. 14(2):131-7 (2007).
2005
Agmon I, Bashan A, Zarivach R, Yonath A. Symmetry at the active site of the ribosome: structural and functional implications. Biol Chem. 386(9):833-44 (2005).
Houben EN, Zarivach R, Oudega B, Luirink J. Early encounters of a nascent membrane protein: specificity and timing of contacts inside and outside the ribosome. J Cell Biol. 170(1):27-35 (2005).
Auerbach-Nevo T, Zarivach R, Peretz M, Yonath A. Reproducible growth of well diffracting ribosomal crystals. Acta Crystallogr D Biol Crystallogr. 61(Pt 6):713-9 (2005).
Pfister P, Corti N, Hobbie S, Bruell C, Zarivach R, Yonath A, Bottger EC. 23S rRNA base pair 2057-2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A->G. Proc Natl Acad Sci USA. 102(14):5180-5 (2005).
2004
Agmon I, Amit M, Auerbach T, Bashan A, Baram D, Bartels H, Berisio R, Greenberg I, Harms J, Hansen HA, Kessler M, Pyetan E, Schluenzen F, Sittner A, Yonath A, Zarivach R. Ribosomal crystallography: a flexible nucleotide anchoring tRNA translocation, facilitates peptide-bond formation, chirality discrimination and antibiotics synergism. FEBS Lett. 567(1):20-6 (2004).
Zarivach R, Bashan A, Berisio R, Harms J, Auerbach T, Schluenzen F, Bartels H, Baram D, Pyetan E, Sittner A, Amit M, Hansen H, Kessler M, Liebe C, Wolff A, Agmon I and Yonath A. Functional aspects of ribosomal architecture: symmetry, chirality and regulation. J. Phys. Org. Chem.. 17: 901–912 (2004).
2003
Bashan A, Zarivach R, Schluenzen F, Agmon I, Harms J, Auerbach T, Baram D, Berisio R, Bartels H, Hansen HA, Fucini P, Wilson D, Peretz M, Kessler M, Yonath A. Ribosomal crystallography: peptide bond formation and its inhibition. Biopolymers. 70(1):19-41 (2003).
Berisio R, Harms J, Schluenzen F, Zarivach R, Hansen HA, Fucini P, Yonath A. Structural insight into the antibiotic action of telithromycin against resistant mutants. J Bacteriol. 185(14):4276-9(2003).
Kleifeld O, Shi SP, Zarivach R, Eisenstein M, Sagi I. The conserved Glu-60 residue in Thermoanaerobacter brockii alcohol dehydrogenase is not essential for catalysis. Protein Sci. 12(3):468-79 (2003).
Ben-Zeev E*, Zarivach R*, Shoham M, Yonath A, Eisenstein M. Prediction of the Structure of the Complex Between the 30S Ribosomal Subunit and Colicin E3 via Weighted-Geometric Docking. J Biomol Struct Dyn. 20(5) 669-76 (2003).
Bashan A*, Agmon I*, Zarivach R, Schluenzen F, Harms J, Bartels H, Franceschi F, Hansen HSA, Kossoy E, Kessler M and Yonath A. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 11(1) 91-102 (2003).
Schluenzen F, Harms J, Franceschi F, Hansen HAS, Bartels H, Zarivach R and Yonath A. Structural basis for the antibiotic activity of ketolides and azalides. Structure11(3) 329-38 (2003).
2002
Henn A, Shi SP, Zarivach R, Ben-Zeev E, Sagi I. The RNA helicase DbpA exhibits a markedly different conformation in the ADP-bound state compared with the ATP- or RNA-bound states. J Biol Chem. 277(48):46559-65. (2002).
Zarivach R*, Ben-Zeev E*, Wu N, Auerbach T, Bashan A, Jakes K, Dickman K, Kosmidis A, Schluenzen F, Yonath A, Eisenstein M and Shoham M. On the Interaction of Colicin E3 with the Ribosome. Biochemie, 447-54 (2002).
Auerbach T., Bashan A., Harms J., Schluenzen F., Zarivach R., Bartels H., Agmon I., Kessler M., Pioletti M., Franceschi F. and Yonath A. Antibiotics Targeting Ribosomes: Crystallographic Studies. Curr Drug Targets – Infectious Disorders, 2, 169-86 (2002)
Harms J., Schluenzen F., Zarivach R., Bashan A., Bartels H., Agmon I. and Yonath A. Protein structure: experimental and theoretical aspects. FEBS Lett, 525, 176-7 (2002).
Zarivach R., Bashan A., Schluenzen F., Harms J., Pioletti M., Franceschi F. and Yonath A. Initiation and inhibition of protein biosynthesis studies at high resolution. Current Protein and Peptide Science, 3, 55-65 (2002).
Vlamis-Gardikas A, Potamitou A, Zarivach R, Hochman A, Holmgren A. Characterization of Escherichia coli null mutants for glutaredoxin 2.
J Biol Chem, 277(13) 10861-8 (2002).
2001
Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Pioletti M, Bartels H, Gluehmann M, Hansen HA, Auerbach T, Franceschi F, Yonath A. High resolution structures of ribosomal subunits: initiation, inhibition and conformational variability. Cold Spring Harb Symp Quant Biol, 66, 43-56 (2001).
Gluehmann M, Zarivach R, Bashan A, Harms J, Schluenzen F, Bartels H, Agmon I, Rosenblum G, Pioletti M, Auerbach T, Avila H, Hansen HA, Franceschi F, Yonath A. Ribosomal crystallography: from poorly diffracting micro-crystals to high resolution structures. Methods, 25, 292302 (2001).
Schluenzen F*, Zarivach R*, Harms J*, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature, 413, 814-21 (2001) .
Harms J*, Schluenzen F*, Zarivach R*, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell, 107, 679-88 (2001) and Supplement.
Pioletti M*, Schluenzen F*, Harms J*, Zarivach R, Gluehmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J, 20, 1829-39 (2001).
2000
Schluenzen F*, Tocilj A*, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A.
Structure of functionally activated small ribosomal subunit at 3.3 A resolution. Cell, 102, 615-23 (2000).

Lorem ipsum dolor sit amet, consectetur adipiscing elit. Ut elit tellus, luctus nec ullamcorper mattis, pulvinar dapibus leo.