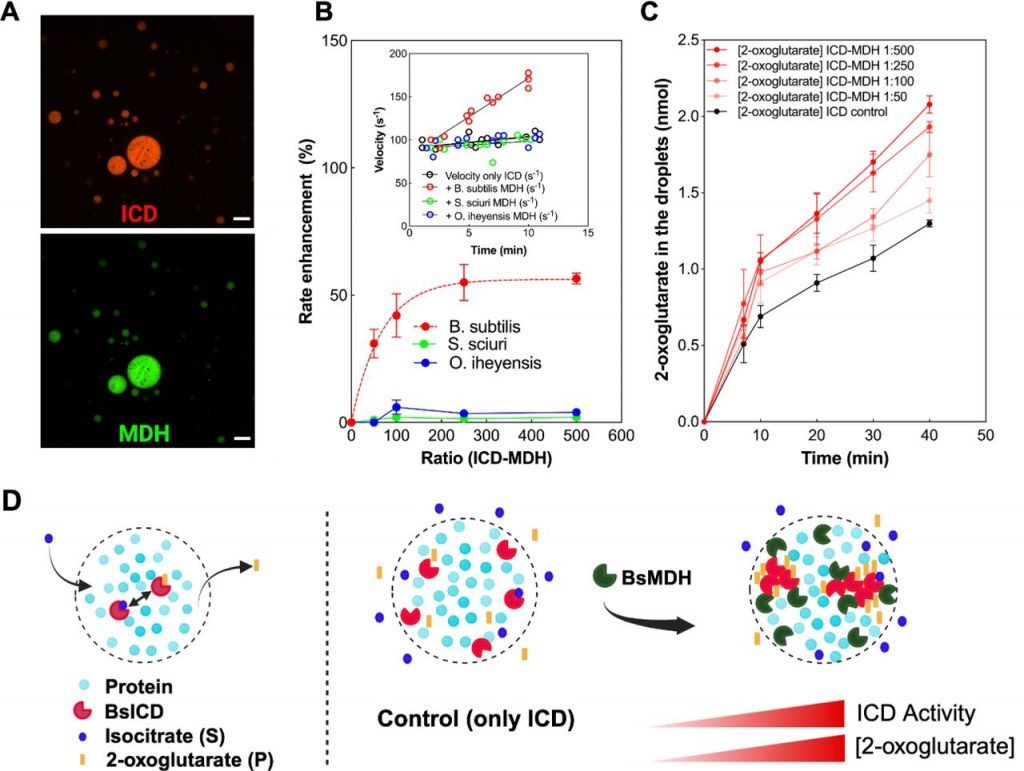
Weronika Jasinska, Mirco Dindo, Sandra M. Correa, Adrian W.R. Serohijos, Paola Laurino, Yariv Brotman, & Shimon Bershtein. Non-consecutive enzyme interactions within TCA cycle supramolecular assembly regulate carbon-nitrogen metabolism. Nat Commun 15, 5285 (2024). https://doi.org/10.1038/s41467-024-49646-7
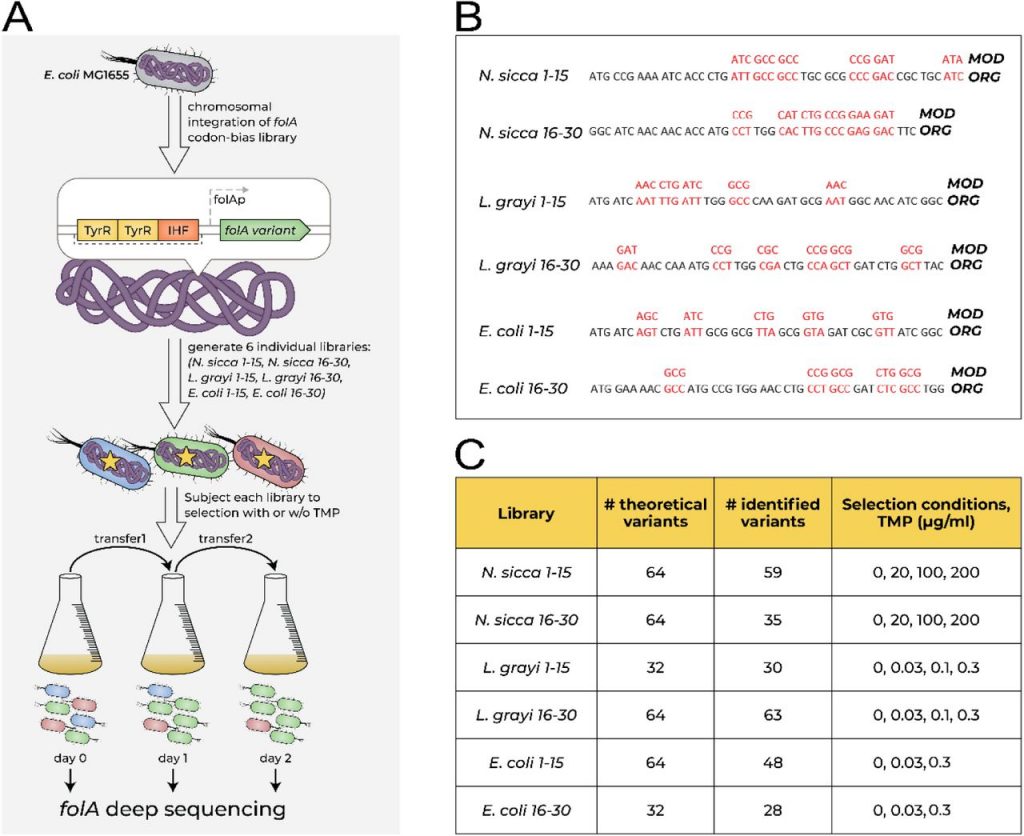
Gencel M, Cofino GM, Hui C, Gauthier L, Tsang D, Philpott D, Ramathan S, Menendez A, Bershtein S, & Serohijos AWR (2023). Intra- and inter-species interactions drive early phases of invasion in mice gut microbiota. Biorxiv. doi: https://doi.org/10.1101/2022.12.30.522336
Shaferman M, Gencel M, Alon N, Alasad K, Rotblat B, Tsang D, Serohijos AWR, Alfonta L, & Bershtein S (2023). The fitness effects of codon composition of the horizontally transferred antibiotic resistance genes intensify at sub-lethal antibiotic levels. Mol Biol Evo. 40(6): ahead of print
Stein D, Slobodnik Z, Tam B, Einav M, Akabayov B, BersteinS, ToiberPI D (2022). 4-phenylbutyric acid-Identity crisis; can it act as a translation inhibitor? Aging Cell. 21:e13738
Simon-Baram H, Roth S Z, Niedermayer C, Huber P, Speck M, Diener J, Richter M & Bershtein S. (2022). A high-throughput continuous spectroscopic assay to measure the activity of natural product methyltransferases. ChemBioChem. 4: e202200162
Kleiner D, Shapiro Tuchman Z Shmulevich F, Zarivach R, Shahar A, Kosloff M, & BershteinPI S. (2022). Evolution of homo-oligomerization of methionine S-adenosyltransferase is replete with structure-function constraints. Protein Sci. 31: e4352 doi:10.1002/pro.4352. https://pubmed.ncbi.nlm.nih.gov/35762725/
Bershtein, S., Kleiner, D. & Mishmar, D. Predicting 3D protein structures in light of evolution. Nat. Ecol. Evol. 5, 1195–1198 (2021). https://www.nature.com/articles/s41559-021-01519-8
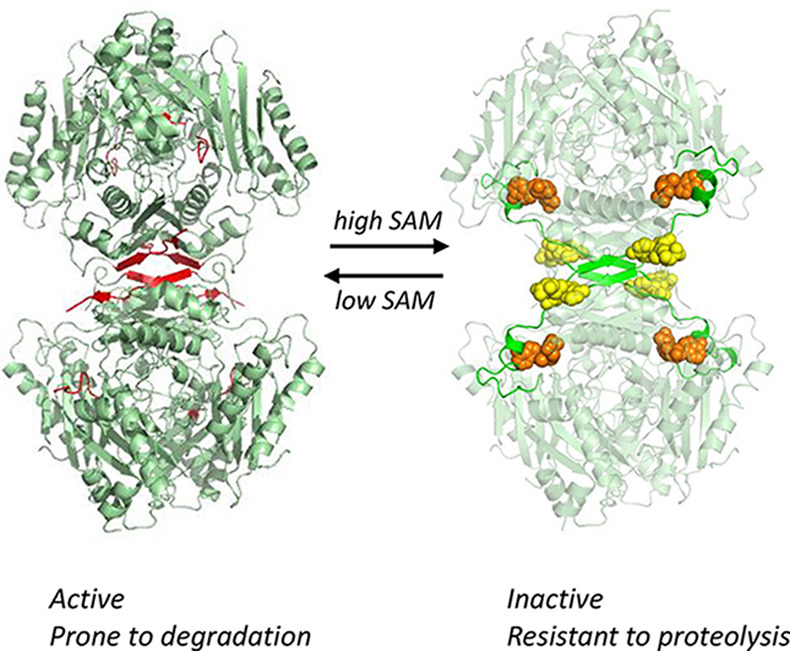
Simon-Baram H, Kleiner D, Shmulevich F, Zarivach R, Zalk R, Tang H, Ding F, & Bershtein S. (2021). SAMase of bacteriophage T3 inactivates E. coli’s methionine S-adenosyltransferase by forming hetero-polymers. mBIO. 12: e01242-21 https://pubmed.ncbi.nlm.nih.gov/34340545/
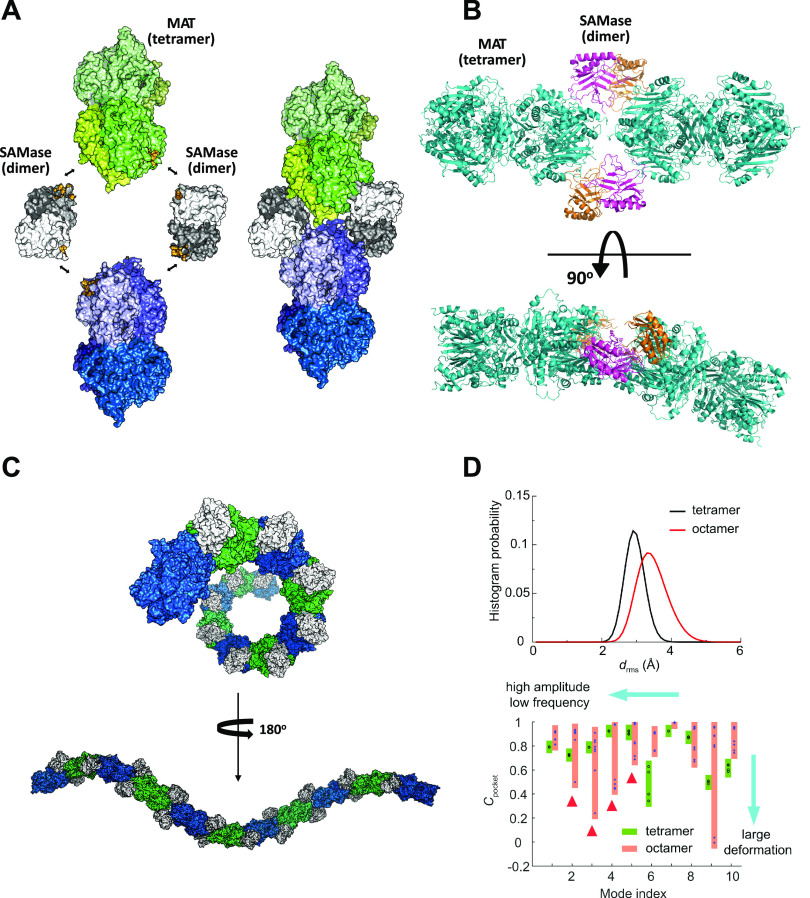
Bhattacharyya, S., Bershtein, S., Adkar, B. V, Woodard, J. & Shakhnovich, E. I. Metabolic response to point mutations reveals principles of modulation of in vivo enzyme activity and phenotype. Mol. Syst. Biol. 17, 1–19 (2021). https://pubmed.ncbi.nlm.nih.gov/34180142/
Galai G, Ben-David H, Levin L, Orth M. F, Grünewald T, Pilosof S, & Bershtein S, & Rotblat, B. (2020) Pan-Cancer Analysis of Mitochondria Chaperone-Client Co-Expression Reveals Chaperone Functional Partitioning. Cancers, 12: 825-838
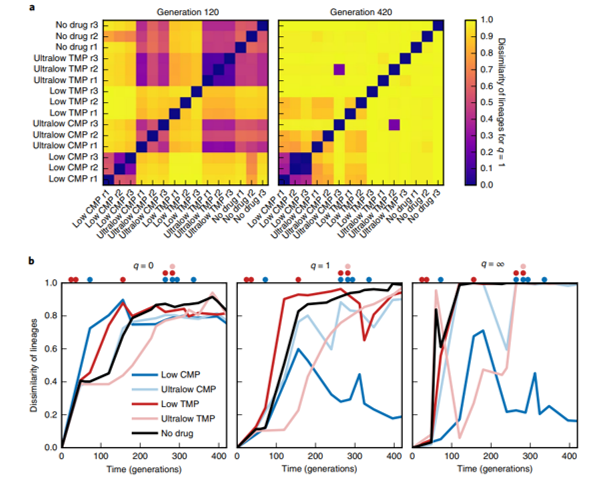
Jasinska W, Manhart M, Lerner J, Gauthier L, Serohijos AWR, B. S. Chromosomal barcoding of E. coli populations reveals lineage diversity dynamics at high resolution. Nat. Ecol. Evol. 4, 437–452 (2020). https://www.nature.com/articles/s41559-020-1103-z
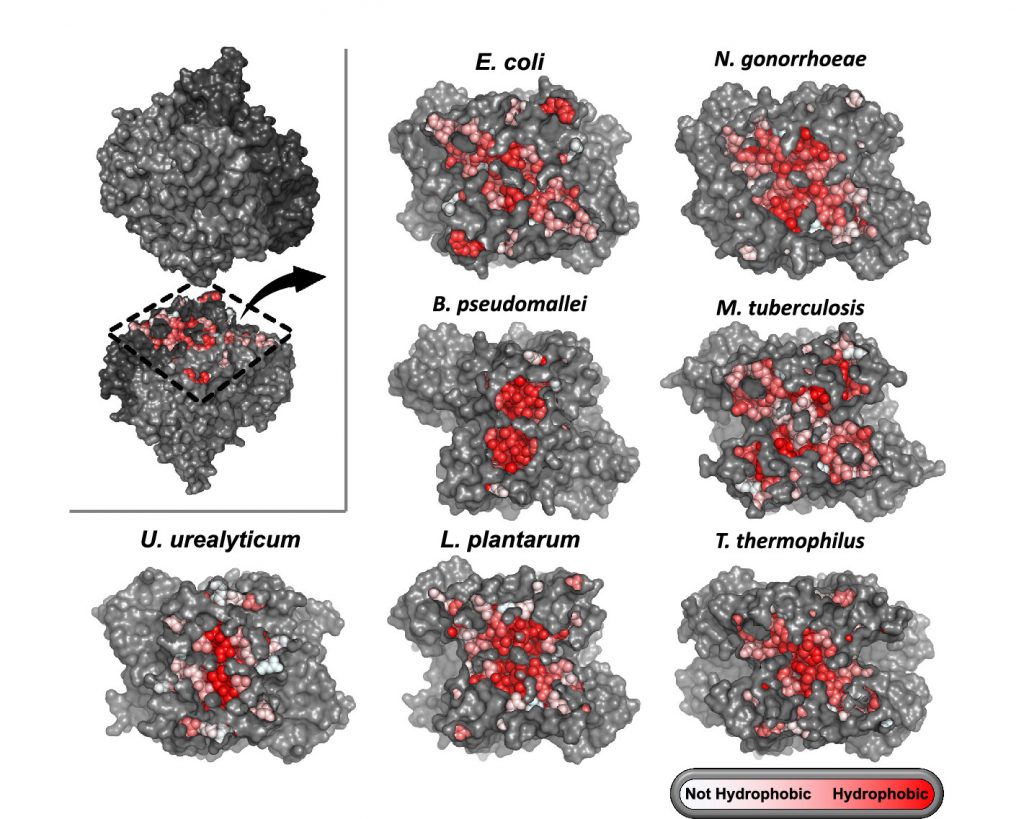
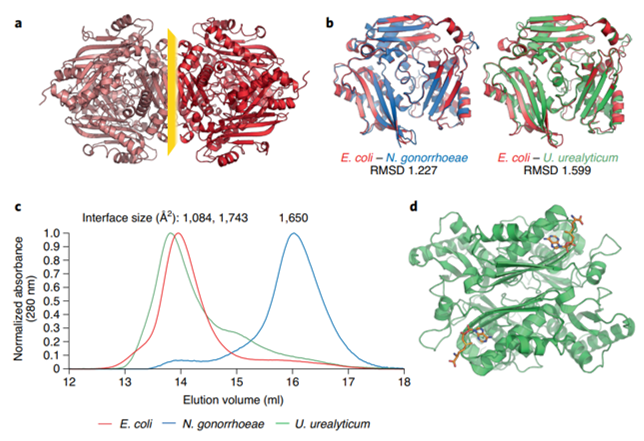
Kleiner D, Shmulevich F, Zarivach R, Shahar A, Sharon M, Ben-Nissan G, Bershtein S. The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase. J Mol Biol. 2019 Dec 6;431(24):4796-4816. doi: 10.1016/j.jmb.2019.09.003. Epub 2019 Sep 12. https://pubmed.ncbi.nlm.nih.gov/31520601/
Bershtein, S., Serohijos, A. W. & Shakhnovich, E. I. Bridging the physical scales in evolutionary biology: from protein sequence space to fitness of organisms and populations. Curr. Opin. Struct. Biol. 42, 31–40 (2017). https://pubmed.ncbi.nlm.nih.gov/27810574/
Bhattacharyya, S., Bershtein, S. & Shakhnovich, E. I. Gene Dosage Experiments in Enterobacteriaceae Using Arabinose-regulated Promoters. Bio-Protocol 7, 1–6 (2017). https://pubmed.ncbi.nlm.nih.gov/29170750/
Bershtein S, Bhattacharyya S, Yan J, Argun T, Gilson AI, Trauger S, & Shakhnovich EI. (2016) Transient protein-protein interactions perturb E. coli metabolome and cause gene dosage toxicity. eLife. pii: e20309 doi: 10.7554/eLife.20309
RodriguesJV, Bershtein S, Li A, Lozovsky E, Hartl DL, & Shakhnovich EI. (2016). Biophysical principles predict fitness landscapes of drug resistance. PNAS. 113(11): E1470-8
Bershtein S, Serohijos AWR, Bhattacharyya S, Manhart M, Choi J-M, Mu W, Zhou J, & Shakhnovich EI. (2015) Protein homeostasis imposes a barrier on functional integration of horizontally transferred genes in bacteria. PloS Genet. 11(10): e1005612.
Palmer AC, Toprak E, Baym M.S, Kim S, Veres A, BershteinC S & KishonyPI R. (2015) Delayed commitment to evolutionary fate in antibiotic resistance fitness landscapes. Nat Commun. 6: 7385-7392.
Bershtein S, Choi J-M, Bhattacharyya S, Budnik B, & Shakhnovich IE. (2015) Systems-level response to point mutations in a core metabolic enzyme modulates genotype-phenotype relationship. Cell Rep. 11: 645-56.
Bershtein S, Mu W, Serohijos A.W, Zhou J, & Shakhnovich IE. (2013) Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Mol Cell. 49: 133-44. 7.
Bershtein S, Mu W & Shakhnovich IE. (2012) Soluble oligomerization provides a beneficial fitness effect on destabilizing mutations. PNAS. 109: 4857-4862.
Bershtein S & Tawfik DS. (2008) Advances in laboratory evolution of enzymes. Curr Opin Chem Biol. 12: 151-158.
Bershtein S & Tawfik DS. (2008) Ohno’s model revisited: Measuring the frequency of potentially adaptive mutations under various mutational drifts. Mol Biol Evol. 25: 2311-2318.
Bershtein S, Goldin K & Tawfik DS. (2008) Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol. 379: 1029-1044
Bershtein S, Segal M, Bekerman R, Tokuriki N & Tawfik DS. (2006) Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein.
Nature. 444: 929-932.
