Molecular and experimental evolution
Our research
SHOW MORE
There is a growing appreciation that horizontal gene transfer (HGT) is a major force in bacterial evolution. We study the molecular barriers that control the accommodation of new genes in the recipient organism using laboratory evolution. In particular, we unravel the dynamics of re-wiring of metabolic and regulatory networks of the host accompanying the evolutionary adaptation upon acquisition of foreign genes.
Bacterial adaptation by interface evolution of methionine S-
adenosyltransferases
SHOW MORE
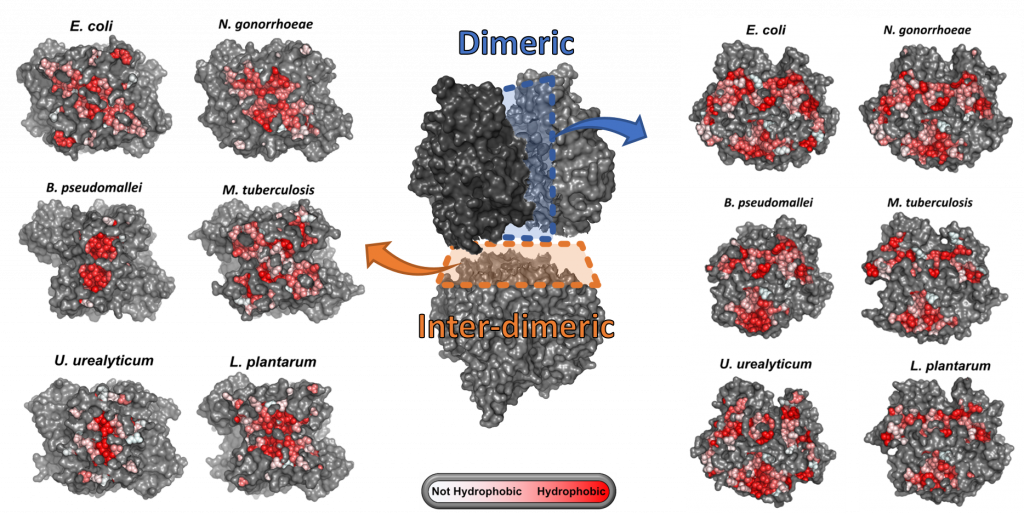
We investigate the molecular mechanisms that drive evolutionary variation in homomeric complexes by addressing the following questions. How protein interface evolution is linked to variation in function and structural integrity of homomeric complexes. How the emerging properties of the homomeric complexes affect organismal fitness and adaptability. We address these questions by focusing on bacterial methionine S-adenosyltransferases (MATs), an essential homo-tetrameric enzyme of central metabolism. integration of both approaches will allow us to establish a genotype-phenotype-fitness map of MAT evolution.
High-resolution lineage tracing of biofilm evolution
SHOW MORE
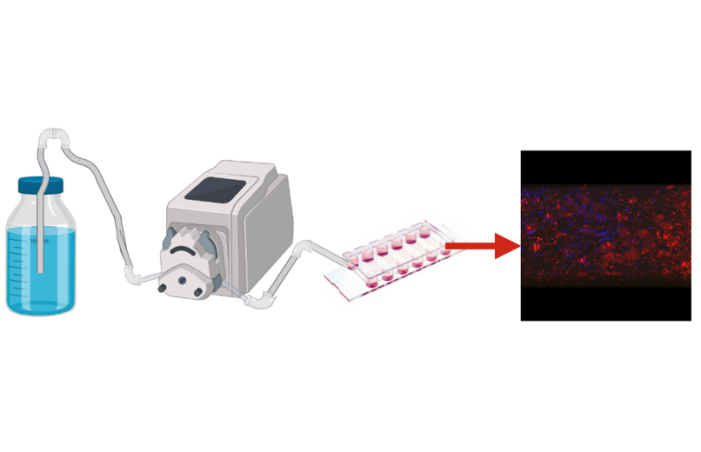
Biofilms are a major form of microbial life in which multiple metabolically diverse bacterial species form densely populated communities. The multi-niche biofilm environment sets a stage for evolution of the inter-species interactions between multiple microbial species inhabiting the biofilm. We study this process by using a high-resolution chromosomal barcoding of the individual cells comprising the mixed-species microbial biofilm community
Understanding the emergence of drug resistance by comparative laboratory evolution empowered by high-resolution lineage-tracing

SHOW MORE
Adaptive evolution, such as evolution towards antibiotic resistance, operates at vastly different scales of biological organization. The molecular and population scales are usually studied independently, hindering our ability to predict the evolutionary paths to antibiotic resistance, and, consequently, to understanding and controlling drug resistance. We apply a powerful experimental approach to study the evolutionary dynamics of drug resistance at a population level. The population scale analysis of the evolving populations relies on chromosomal barcoding with unique random DNA sequences that can be tracked over time.

Functional coordination of the metabolic and protein-protein interaction networks via metabolon formation
SHOW MORE
It is well established that enzymes of central metabolism tend to assemble into transient supramolecular complexes. We investigate the functional significance of the interactions within these complexes, particularly between enzymes catalyzing non-consecutive reactions.
Our contacts
Contact us via email: shimonb@bgu.ac.il Office (+972) 8 642 8808, Fax (+972) 8 642 8796 Ben-Gurion University of the Negev, Building 40, Room 302

