Our toolbox is composed of:
Quantitative live cell imaging:
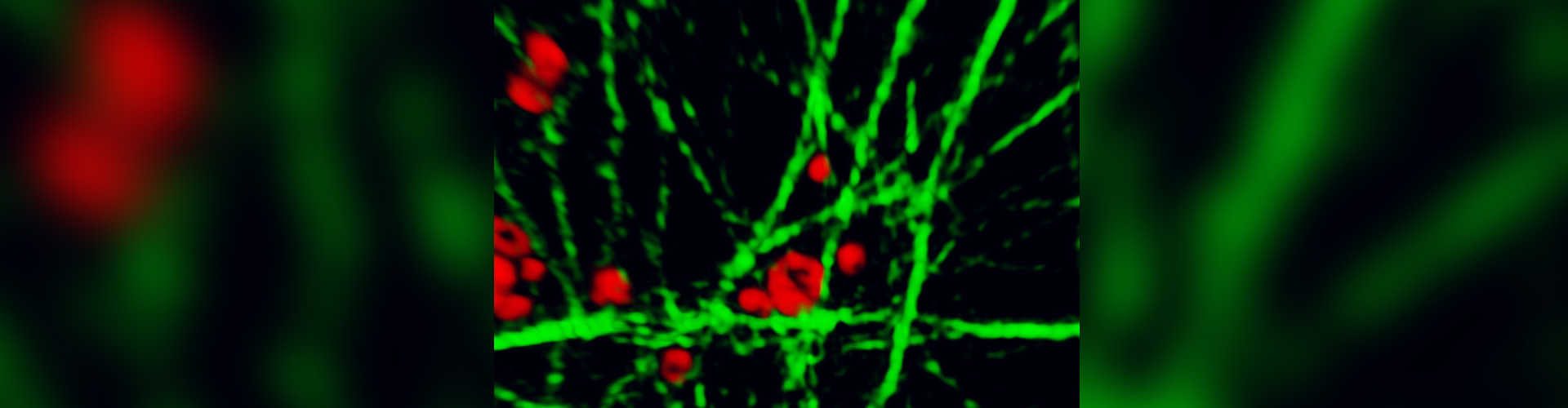
Our lab owns a Marianas Spinning Disk confocal system (3I) equipped with an Evolve EMCCD camera and a Flash4 sCMOS camera, the vector system for photomanipulation and a full incubation system. This system provides the high imaging speed, high sensitivity and high dynamic range required to measure protein dynamics in cells at close to physiological conditions. We use this system to capture the dynamics of abscission and of proteins in abscission in WT and mutant cells.
Super Resolution Microscopy
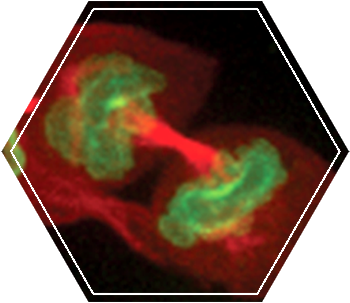
We use Structured Illumination Microscopy (SIM) and Stochastic Optical Reconstruction Microscopy (STORM) to visualize the organization of ESCRTs in cells. Our lab is equipped with the ELYRA PS.1 system (Zeiss) capable of SIM and STORM imaging.
Structured Illumination Microscopy (SIM):
As Dr. Mike Davidson from Florida State University said at the 2010 ASCB meeting in Philadelphia: “SIM is the future of super resolution microscopy technique”. Although the resolution increase in SIM is only double (X100nm, X100nm, X350 nm) it has a few advantages over other super resolution techniques making it the most attractive technique for cell biology studies. SIM is Applicable to most biological samples (up to 10 micron thick) and can be used with any fluorescent probe (antibodies or fluorescent proteins). We routinely use this technique to visualize the organization of different ESCRT components under different cellular conditions.
More information on the principles of SIM can be found here.
Stochastic Optical Reconstruction Microscopy (STORM)
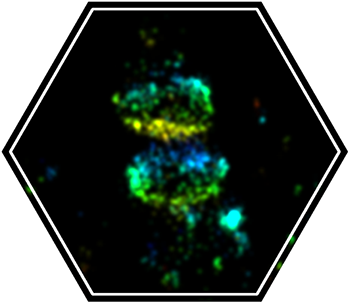
STORM is a microscopy technique capable of achieving 30 nm lateral resolution. This resolution is achieved by stochastically turning “on” a small subset of the fluorescent molecules and resolving their point spread function (PSF). STORM relies on the ability fluorescent molecules to be turned “on” and “off” and therefore only a subset of fluorephores are suitable for this application. This process is repeated for many cycles until the entire population of fluorophores is mapped. The superb resolution of STORM allows resolving the structural organization of macromolecular complexes in cells at the nanometer scale. We use STORM to resolve the structural organization of the ESCRT complex in cells.
More information on the principles of SIM can be found here.

Cryo-Soft-X-Ray tomography
Cryo-Soft-X-Ray tomography (cryo-SXT) is a complementary technique to EM. In cryo-SXT, the ultrastructeral organization of cellular components can be resolved with ~30 nm resolution inside cells. The penetration depth of cryo-SXT is up to 10 micrometers meaning that the entire cell can be imaged intact, without sectioning. The technique is compatibly with tomography providing a 3D view of the cell. In SIM, the sample is cryo-fixed and subjected to low radiation damage insuring an accurate documentation of the native state of the cellular complex.
We adapted this approach to visualize the structural organization of the intercellular bridge during abscission using a correlative light-X-ray approach. Using this approach, we resolved the organization of cortical filaments that reside at the intercellular bridge at different stages of abscission.
We currently use this approach to study the contribution of different ESCRT components to the structure of these filaments. Cryo-SXT experiments are carried out at the synchrotron station in Berlin (BESSY-II). This work is performed in collaboration with Michael Elbaum (WIS).