
ESCRTs in animal cell division
At the end of mammalian cell division, the two nascent daughter cells are connected by a narrow membrane tube called the intercellular bridge. Cutting of the intercellular bridge, termed abscission, is driven by the ESCRT complex. The spatiotemporal characteristics of abscission are ideal for high-end microscopy techniques. To elucidate the mechanism of ESCRTs in physiological context we therefore apply high-resolution microscopy techniques to visualize ESCRTs in mammalian cell abscission. To obtain an inclusive understanding of ESCRT function in abscission, we visualize abscission in both mammalian tissue culture cells and in a developmental system (zebrafish embryogenesis). By combining information obtained from a variety of microscopy tools including live cell imaging, Cryo-tomography and Super resolution microscopy we generate mechanistic models for ESCRT-mediated membrane abscission.
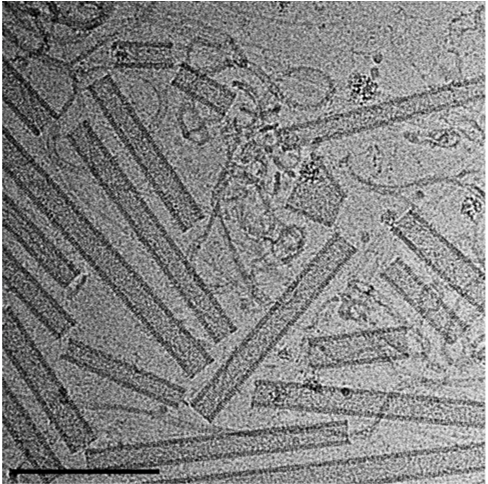
Function of ancient ESCRT systems
The origin of the eukaryotic cell 1.5-2 billion years ago is one of the most profound transitions in the evolutionary history of life. Unlike prokaryotes (Bacteria and Archaea), eukaryotic cells are composed of membrane bound organelles that communicate with one another via membrane carriers. Therefore, studying membrane shaping proteins in ancient lifeforms can get us closer into understanding the membrane-based processes that occurred through evolution. Specifically, we study the ESCRT complex encoded in the recently identified Asgard archaea, which is considered to be the closest prokaryotic ancestor of eukaryotes. By combining in vitro biochemical studies and collaborating with leading groups in the archaea field we aim to shed light on the membrane remodeling capabilities that enabled eukaryogenesis.

Our tool box
Over the past decade, we have developed a comprehensive toolbox to explore and resolve the structural organization of ESCRT complexes in vitro and within cellular environments. Over the past decade, our efforts have focused on building a microscopy-based approach to visualize the ESCRT machinery across various scales. This includes visualizing ESCRT polymers in vitro, as well as unraveling their organization and dynamics in mammalian tissue culture cells and living embryos.
To achieve this, we integrate advanced techniques such as live-cell imaging, super-resolution microscopy, and cryo-electron tomography, enabling us to capture detailed insights into ESCRT complexes at multiple levels of resolution.





