
In vivo monitoring of DNA replication, transcription dynamics and DNA damage
DNA replication is a highly complex process, carried out accurately and efficiently by all living organisms. During this process, the replication machinery must work together with a variety of related cellular processes, such as transcription, DNA repair, chromatin packaging and the cohesion of sister chromatids. In addition, replication must proceed efficiently through a variety of problematic DNA sequences including triplet repeat sequences and G quadruplex (G4) structures.
To obtain mechanistic insights into replication, transcription and DNA damage in living cells, we developed a live-cell microscopy approach for monitoring how single replisomes progress through highly transcribed genes, damaged DNA, tandem repeats or G4 sequences, at a defined genomic locus. By site-specific fluorescent labelling of gene transcription or DNA damage sites, our approach provides unique mechanistic insights into the complex interplay between DNA replication, transcription dynamics or DNA repair processes.
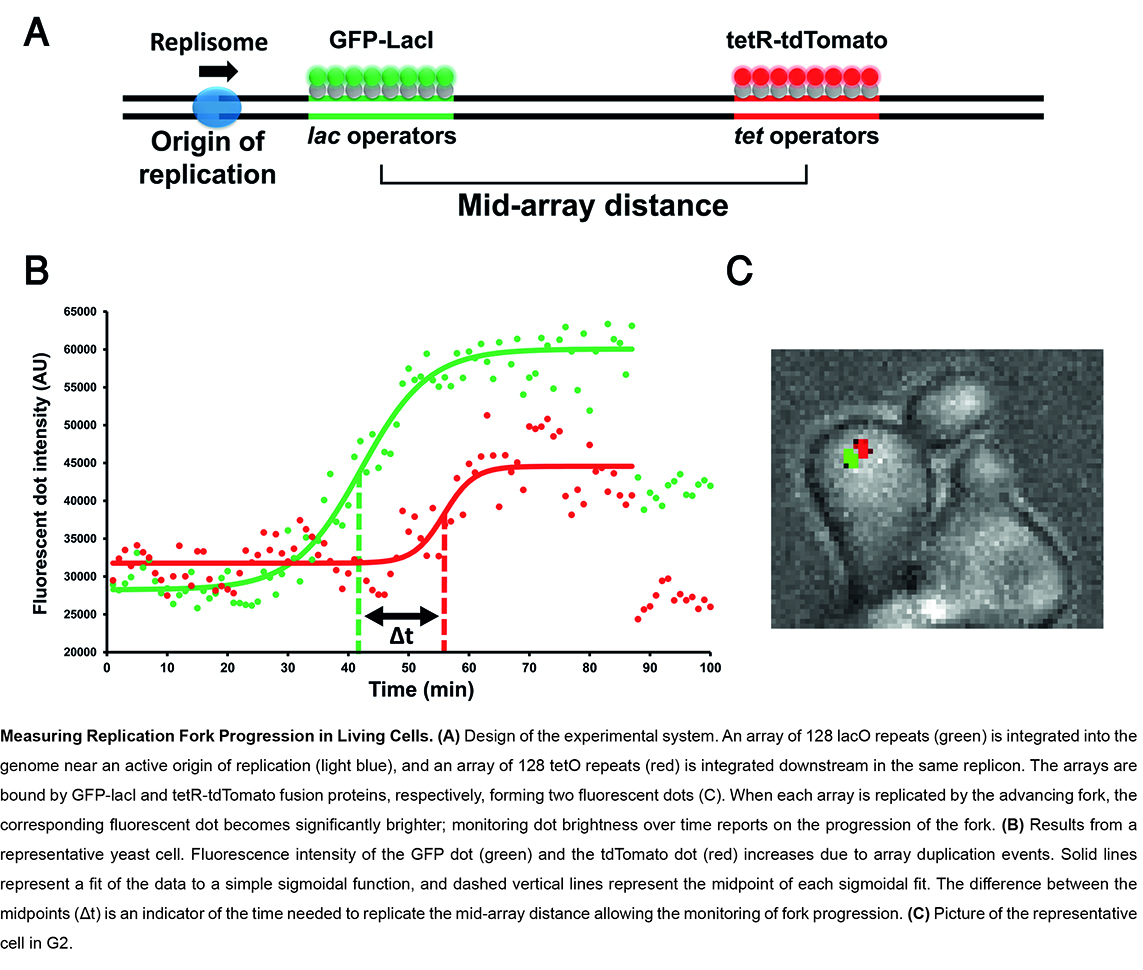
We focus on three main research directions:
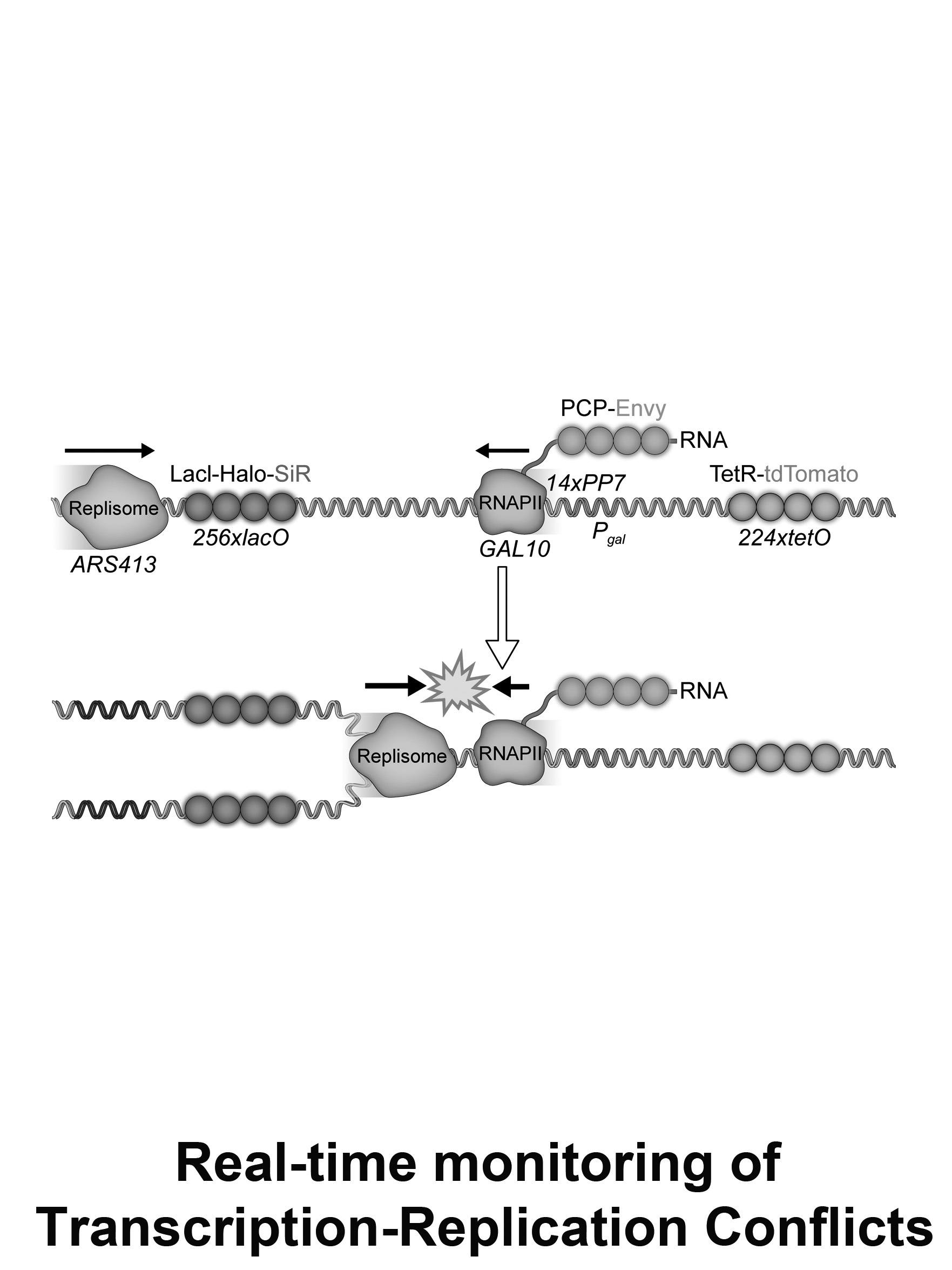
Real-time monitoring of Transcription-Replication Conflicts
The coexistence of DNA replication and transcription during S-phase requires their tight coordination to prevent harmful conflicts. Our lab developed a live-cell imaging approach for the real-time monitoring of replisome progression and transcription dynamics, at the same locus, during a transcription-replication encounter. We study the mechanisms that enable transcription-replication coordination focusing on R-loop formation.
We address on the following questions:
- How active transcription affects replisome progression through the gene?
- What is the transcription dynamic prior, during and following gene duplication?
- What are the effects of transcription-replication encounters during conflicts?
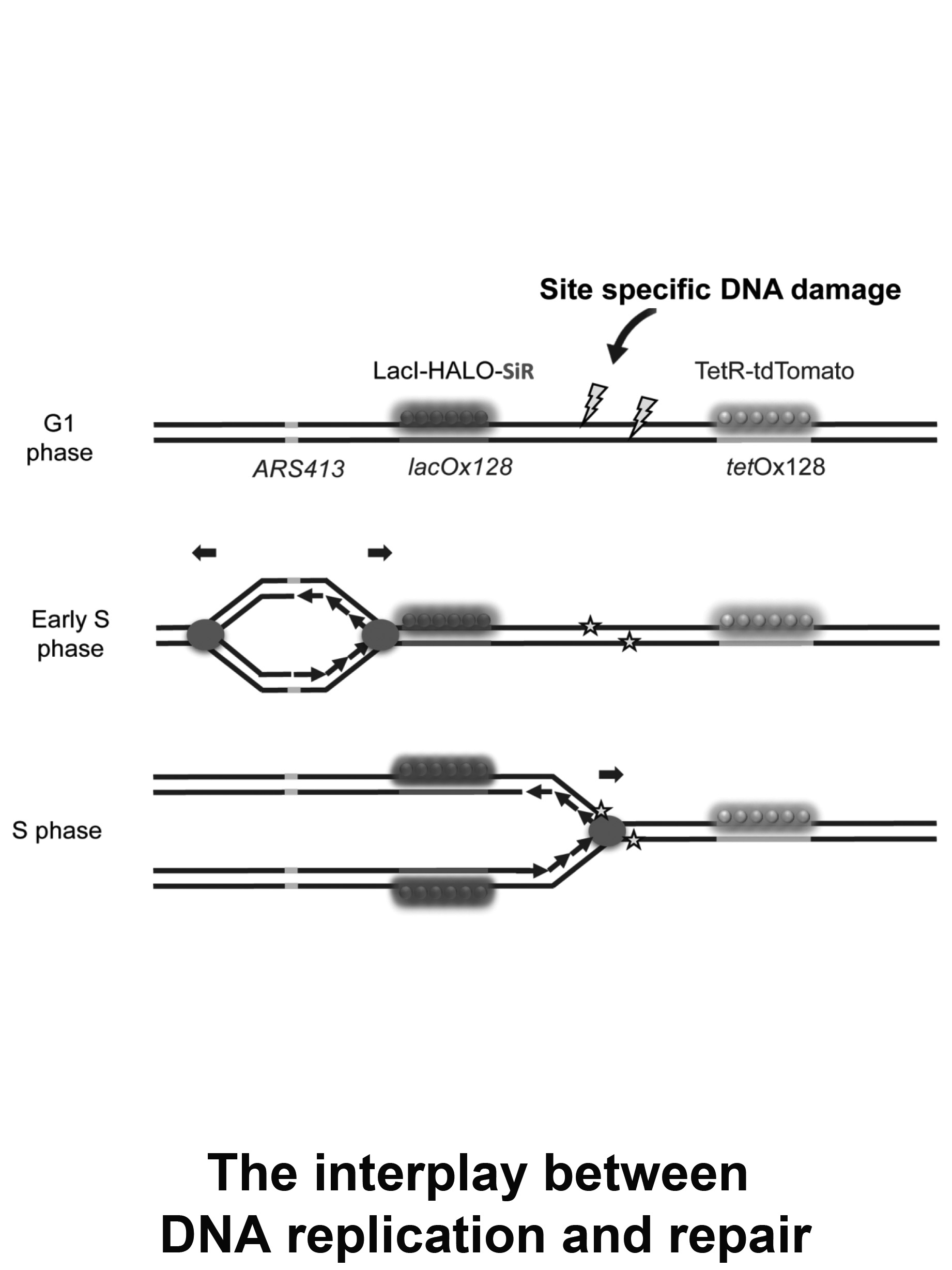
The interplay between DNA replication and repair
DNA damage is extremely abundant due to the constant exposure of cells to UV irradiation or carcinogens. A variety of pathways have evolved to repair damaged DNA and minimize the level of mutagenesis. Our lab developed an approach for the real-time monitoring of replication fork progression through site-specific DNA damage. This approach allows us to reveal mechanistic insights regarding the interplay between replication fork progression through damaged DNA and repair processes.
We focus on the following questions:
- What is the extent of replication slowdown at damaged DNA?
- How different repair pathways affect fork progression through damaged DNA?
- What are the dynamics of DNA damage bypass and repair during replication?
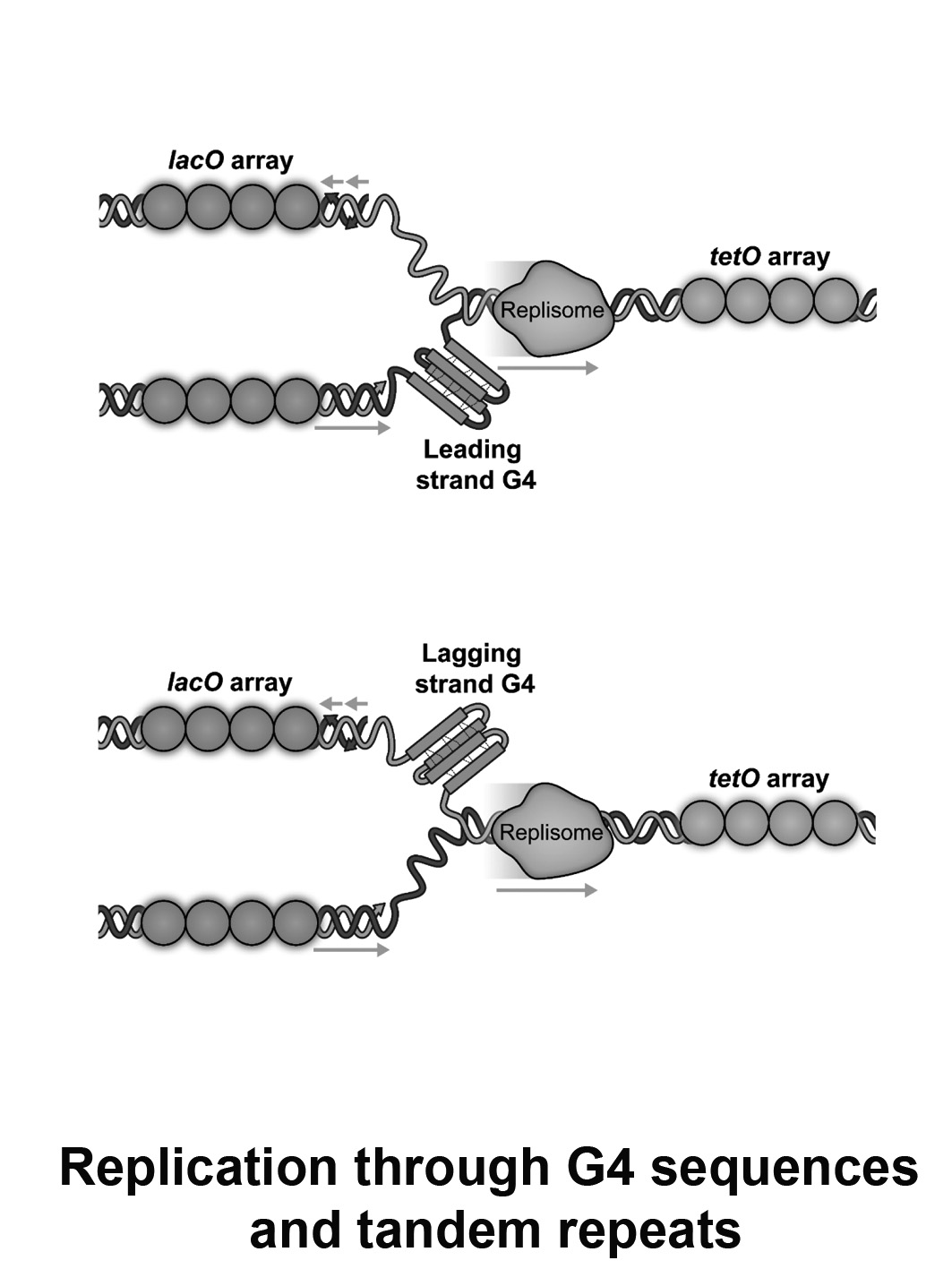
Replication through G4 sequences or tandem repeats
G quadruplex (G4) structures and trinucleotide repeats such as (GAA)n are a source of genetic instability and can lead to many genetic disorders. Genetic defects including contractions, expansions and mutagenesis at these sequences take place during DNA replication. Our lab developed a live-cell imaging approach to quantitatively measure the progression rates of single replication forks through tandem trinucleotide repeats or G4 sequences in individual yeast cells.
Using this approach we address the following questions:
- What are the mechanisms that facilitate fast replication through these sequences?
- What is the relation between replication fork slow down and genetic instability?
- What are the repair factors that are involved in replication through these sequences?